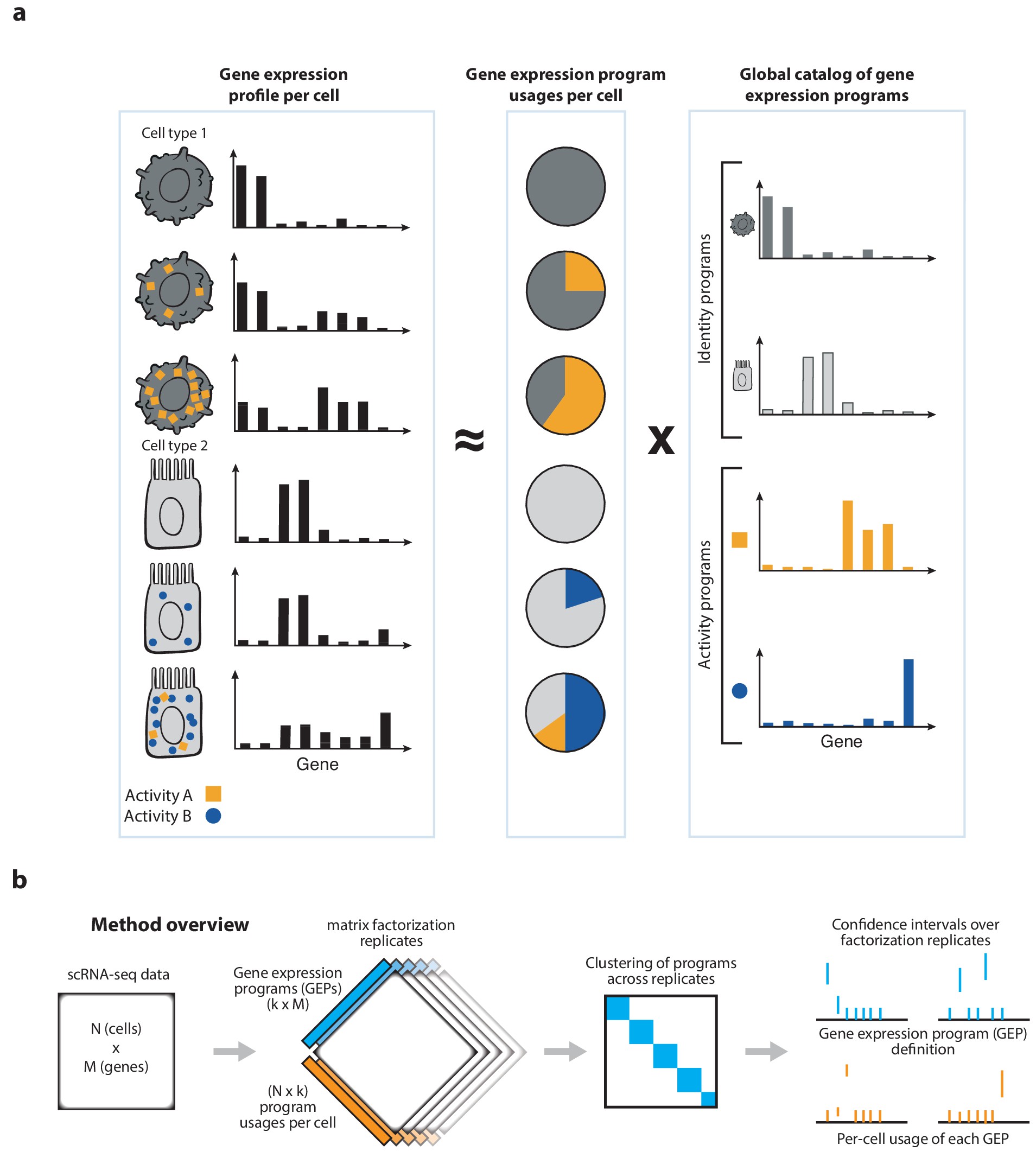
cNMF is an analysis pipeline for inferring gene expression programs from single-cell RNA-Seq (scRNA-Seq) data.
It takes a count matrix (N cells X G genes) as input and produces a (K x G) matrix of gene expression programs (GEPs) and a (N x K) matrix specifying the usage of each program for each cell in the data. You can read more about the method in the publication [here] and check out examples on simulated data and PBMCs.
cNMF-Snakemake is a Snakemake pipeline for running cNMF. This is still in development and is not ready for use OOB as it stands. The code is functional but proper examples have not been set up yet and some bugs need ironing out.
Thank you to Dylan Kotliar for developing cNMF, it has proved very useful for my analyses.