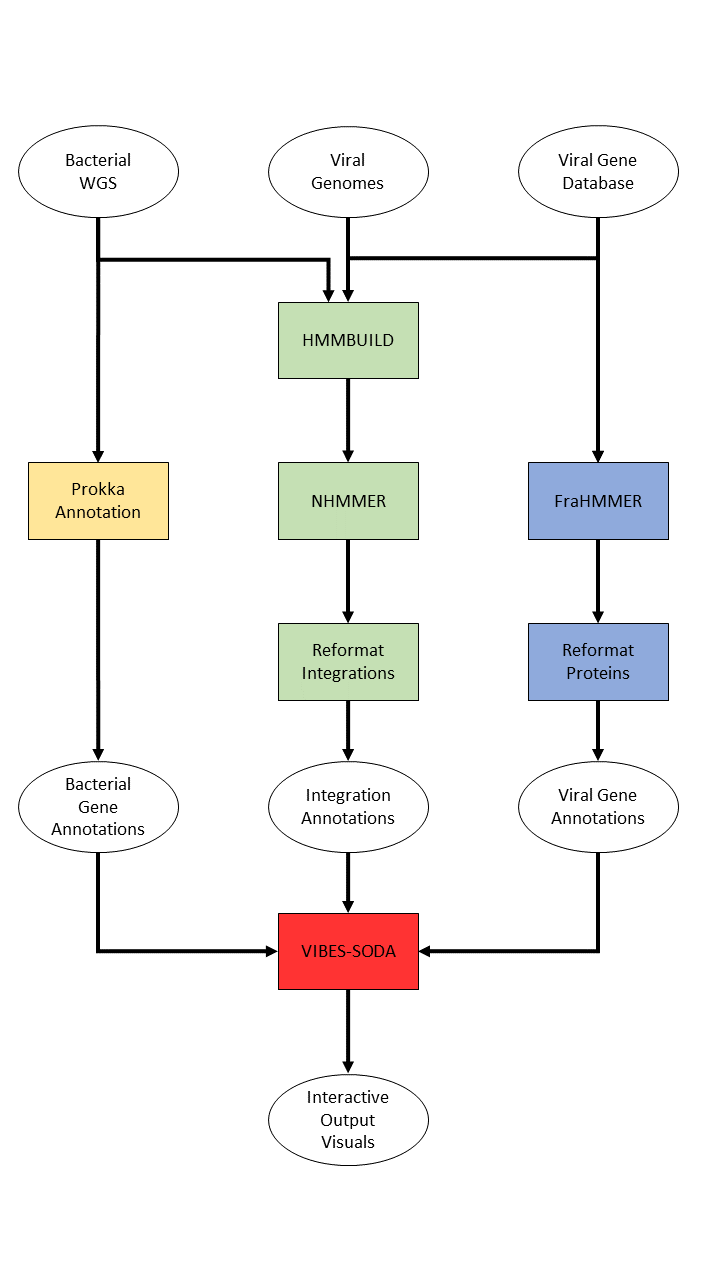
VIBES (Viral Integrations in Bacterial gEnomeS) is a Nextflow-based automated sequence similarity search pipeline that can search for prophage integrations in bacterial whole genome sequence, annotate bacterial and viral proteins, and produce interactive HTML visual output. Users provide VIBES with bacterial whole genome sequence to search in and prophage genomes to search for.
- Workflow automation with Nextflow
- Dependency management via VIBES docker image
- Prophage integration annotation with nhmmer
- Bacterial gene annotation with Prokka
- Viral gene annotation with FraHMMER and PHROGS
- Output visualization with VIBES-SODA
-
Install Java 11 or later, if necessary
-
Install Nextflow, if necessary
-
Install your preferred container management software. Any one of these three should work (or you could install other software that can execute Docker images):
- Docker
- Singularity
- Apptainer
- Only one of the above needs to be installed to run VIBES via its Docker image
-
Install git, if necessary
-
Clone the VIBES GitHub repo to your desired destination:
git clone https://github.com/TravisWheelerLab/VIBES.git -
Navigate to the default viral gene database directory and download:
cd VIBES/nextflow_workflow/resources/db/wget https://zenodo.org/records/10372885/files/phrog_v4.bathmm -
To run VIBES, enter the
VIBES/nextflow_workflow/directory:cd VIBES/nextflow_workflow
- Install VIBES dependencies:
- Python 3 (v3.8.10)
- Perl (v5.30.0)
- nhmmer (v3.3) and Easel (v0.48)
- FraHMMER (v?)
- Prokka (v1.14.6)
-
Install Java 11 or later, if necessary
-
Install Nextflow, if necessary
-
Install git, if necessary
-
Clone the VIBES GitHub repo to your desired destination:
-
Navigate to the default viral gene database directory and download:
cd VIBES/nextflow_workflow/resources/db/wget https://zenodo.org/records/10372885/files/phrog_v4.bathmm -
To run VIBES, enter the
VIBES/nextflow_workflow/directory:cd VIBES/nextflow_workflow
The VIBES workflow is managed by Nextflow and run via the nextflow run command. To invoke VIBES with nextflow run, the user must supply three necessary arguments:
- A nextflow workflow file, in this case
workflow.nf(found inVIBES/nextflow_workflow), that defines the VIBES workflow - A parameters file in .YAML format, which will contain values that might change from run to run (such as input file locations, where to store output, and which parts of the pipeline should be run)
- A profile described in
nextflow.configthat specifies how Nextflow should run VIBES on your compute environment (Are you on an HPC managed by SLURM? Running VIBES locally on a laptop/desktop?)
These three components are supplied to Nextflow as follows: nextflow run workflow.nf -params-file your_params.yaml -profile your_profile. In general, your_params.yaml will be modified from run to run, your_profile will likely only be modified when you set VIBES up on a new compute system, and workflow.nf will be modified only when advanced users want to change how the VIBES workflow operates. The basics of setting up a parameters YAML file and a Nextflow profile are explored below.
The parameters file contains values that might change from run to run: Where to gather input files from, where output should be stored, input sequence type (dna/rna/amino), and which parts of the pipeline should be run. An example parameters file is provided with VIBES at VIBES/nextflow_workflow/fixture_params.yaml and includes all fields VIBES expects parameters files to provide.
Parameters files should be in YAML format, in which a variable is followed by a colon and then a value. For example, in this line genome_files: ${projectDir}/../fixtures/5_full_bac_2_vir/*.fna from fixture_params.yaml, genome_files is the variable name and ${projectDir}/../fixtures/5_full_bac_2_vir/*.fna is the value assigned to the variable. Changing variable names will result in VIBES crashing, so only values should be changed unless the user also modifies workflow.nf.
In Nextflow parameters files, some environment variables can be accessed that can make it easier to reference files outside of the directory that Nextflow is being run in. ${projectDir}, for instance, points Nextflow to the directory that workflow.nf is located in (in this case, VIBES/nextflow_workflow/). Using these environment variables to point to file path(s) is important since Nextflow runs each portion of the workflow from subdirectories in a work/ directory, so relative file paths will not point to the correct locations unless they inlcude ${projectDir} or ${launchDir}.
Profiles tell Nextflow how it should try to run tasks in the workflow: should it use Docker or not? Should it submit jobs to a scheduler (i.e. SLURM) or run them itself? Essentially, profiles set default options for tasks across the VIBES workflow that hold unless specifically overwritten in a task definition. VIBES/nextflow_workflow/nextflow.config contains example profiles for the VIBES workflow and is where users should store their own profiles as needed.
A couple of key profile options to pay attention to: process.executor allows users to set whether Nextflow should submit tasks to an executor like SLURM (list of job managers supported by Nextflow)) or run the tasks locally. Similarly, docker.enabled and process.container tell Nextflow to use Docker to run containers and sets the default container for all tasks to the value of process.container. process.clusterOptions will be an important profile setting for most users who run VIBES on a HPC environment- this setting appends its value to all commands submitted to a job manager (i.e. SLURM) and is a useful place to set cluster options such as partition or an account to be billed.
Full profile documentation can be found here.
Once you've created a YAML file with your preferred parameters and a profile for your environment, you can launch VIBES with nextflow run workflow.nf -params-file your_params.yaml -profile your_profile. If you want to resume a run of VIBES after resolving whatever issue stopped it, you can resume the previous run by adding -resume to your nextflow run command (nextflow run workflow.nf -params-file your_params.yaml -profile your_profile -resume).
To verify that the workflow is working, we've included a test case in test_input_output/test:
From nextflow_workflow, run the following command and compare output to expected_output:
nextflow run workflow.nf -params-file ../test_input_output/test/test.yaml -profile local_docker
Parameters files are YAML format files containing information such as the location of input bacterial genome sequence, input prophage genome sequences, and which parts of the VIBES pipeline should be run. In YAML format, a variable is followed by a colon and then a value. For example, in this line genome_files: ${projectDir}/../fixtures/5_full_bac_2_vir/*.fna from fixture_params.yaml, genome_files is the variable name and ${projectDir}/../fixtures/5_full_bac_2_vir/*.fna is the value assigned to the variable. Changing variable names will result in VIBES crashing, so only values should be changed unless the user also modifies workflow.nf.
In the example parameter file VIBES/nextflow_workflow/fixture_params.yaml, some values are preceeded by ${projectDir}. This is a Nextflow environment variable that tells the workflow to start from whichever directory the workflow file lives in (by default, VIBES/nextflow_workflow/) when following a file path. We recommend using ${projectDir} as the root for relative paths to input and output files or directories.
A complete list of VIBES workflow parameters in the YAML file:
- Basic options:
- genome_files: Path to input bacterial genome sequences in FASTA format. Using glob patterns (i.e.
*.fasta) allows for multiple matching files to be selected (more information on glob patterns) - phage_file: Path to input FASTA file containing all phage genomes you want to search for.
- phage_seq_type: dna/rna/amino, depending on phage sequence residues
- output_path: Path to directory where VIBES will save all output data
- genome_files: Path to input bacterial genome sequences in FASTA format. Using glob patterns (i.e.
- Workflow function options:
- detect_integrations: Run the portion of the pipeline that searches bacterial genomes for prophage integrations
- annotate_phage_genes: Annotate proteins on user-provided prophage genomes
- prokka_annotation: Annotate genes on bacterial genomes with Prokka
- zip_prokka_output: Compress output as .tar.gz files to save space
- Prophage gene annotation options:
- viral_protein_db: Path to prophage gene database, which must be in .hmm or .frahmm format
- viral_protein_annotation_tsv: Path to .tsv file with two fields: protein ID and function description, separated by a tab character
In Nextflow parameters files, some environment variables can be accessed that can make it easier to reference files outside of the directory that Nextflow is being run in. ${projectDir}, for instance, points Nextflow to the directory that workflow.nf is located in (in this case, VIBES/nextflow_workflow/). Using these environment variables to point to file path(s) is important since Nextflow runs each portion of the workflow from subdirectories in a work/ directory, so relative file paths will not point to the correct locations unless they inlcude ${projectDir} or ${launchDir}.
nextflow run workflow.nf -params-file your_params.yaml -profile your_profile is the minimum necessary command to launch VIBES, but there are some other useful options worth knowing about:
nextflow -log log_file.log run ...will save a Nextflow log filenextflow run workflow.nf -w /path/to/some/dir/ ...allows users to specify a work directory other thanVIBES/nextflow_workflow/work/, where Nextflow stores the workflow cachenextflow run workflow.nf -with-report report_name.html ...will generate an HTML report of pipeline resource usage after the workflow successfully completes.nextflow run workflow.nf -resume ...instructs Nextflow to pick up where the last run of the pipeline left off, where possible. Allows restarting a crashed pipeline while retaining as much work as possible from the previous run.- For a list of all
nextflow runoptions, and information on other Nextflow command line utilities, see the Nextflow docs
Here, we inclue some example nextflow.config compute profiles. All of these examples are based heavily on the profiles I use to run the workflow. The local profiles should work for any local execution (at least on Unix) and the HPC profiles should work if the correct executor, container management software, and partition and account details are set up. Note that --clusterOptions will depend heavily on your particular system.
This is the simplest case for a profile. It instructs Nextflow to run all operations locally, as local hardware resources allow:
profiles {
local {
// Comments look like this! Here, we set the executor (what Nextflow submits operations to)
process.executor = 'local'
}
}
Similar to the above case, but instructs Nextflow to run all operations inside of a Docker container. Note that this is equivalent to setting a default Docker container for the pipeline, and can be overwritten on a per-process basis in workflow.nf.
profiles {
local_docker {
process.executor = 'local'
process.container = 'connercopeland/vibes-test-frahmmer:latest'
docker.enabled = true // this tells Nextflow to use Docker specifically to execute the container
params.programs_path = '/programs/' // this line should be deprecated
}
}
This example shows how multiple profiles can be stored in nextflow.config and how profiles can be set up to operate on HPC systems.
profiles {
gscc {
process.executor = 'slurm' // here, we tell Nextflow to submit operations via SLURM, rather than to run locally
process.clusterOptions = '--partition=list_of_partitions' // you can use this field to provide options like accounts to bill, partitions to use, etc
}
ua_hpc {
process.executor = 'slurm'
process.clusterOptions = '--partition=standard --account=account --ntasks=1'
process.container = 'connercopeland/vibes-test-frahmmer:latest'
singularity.enabled = true // Here we specify to run the container with Singularity, which is more popular on HPCs
process.scratch = true // ask nextflow to store intermediate fies on nodes instead of in /home, improving performance and reducing I/O
process.cache = 'deep' // sets Nextflow to cache based on input file contents, rather than input file path and date
}
}
Coming Soon
- Thanks for helping make VIBES happen!
- George Lesica
- Jeremiah Gaiser