v0.15 30aug2023
A Stata package to estimate covariate interactions and subgroup-specific treatment effects in meta-analysis
Meta-analysis is a statistical technique for combining results from multiple independent studies, with the aim of estimating a single overall effect. However, it can also be used for assessing how treatment effects vary across participant subgroups. Ideally, this assessment will be based on treatment-covariate interaction effects derived within each trial separately, so that the pooled effects are free from aggregation bias. In particular, metafloat provides a simple way of estimating pooled interactions and "floating" subgroup-specific treatment effects from a set of observed (published, or otherwise pre-aggregated) treatment effects by trial and by subgroup.
See Godolphin et al (2023) for more information on the underlying methodology. The lead author, Dr Godolphin, also gave a Cochrane webinar on the methodology in January 2024.
To install the package directly from GitHub, type from within Stata:
. net describe metafloat, from("https://raw.githubusercontent.com/UCL/metafloat/master/src/")
Currently, documentation on usage and options may be found in the documentation files within Stata. After installation, type in Stata:
. help metafloat
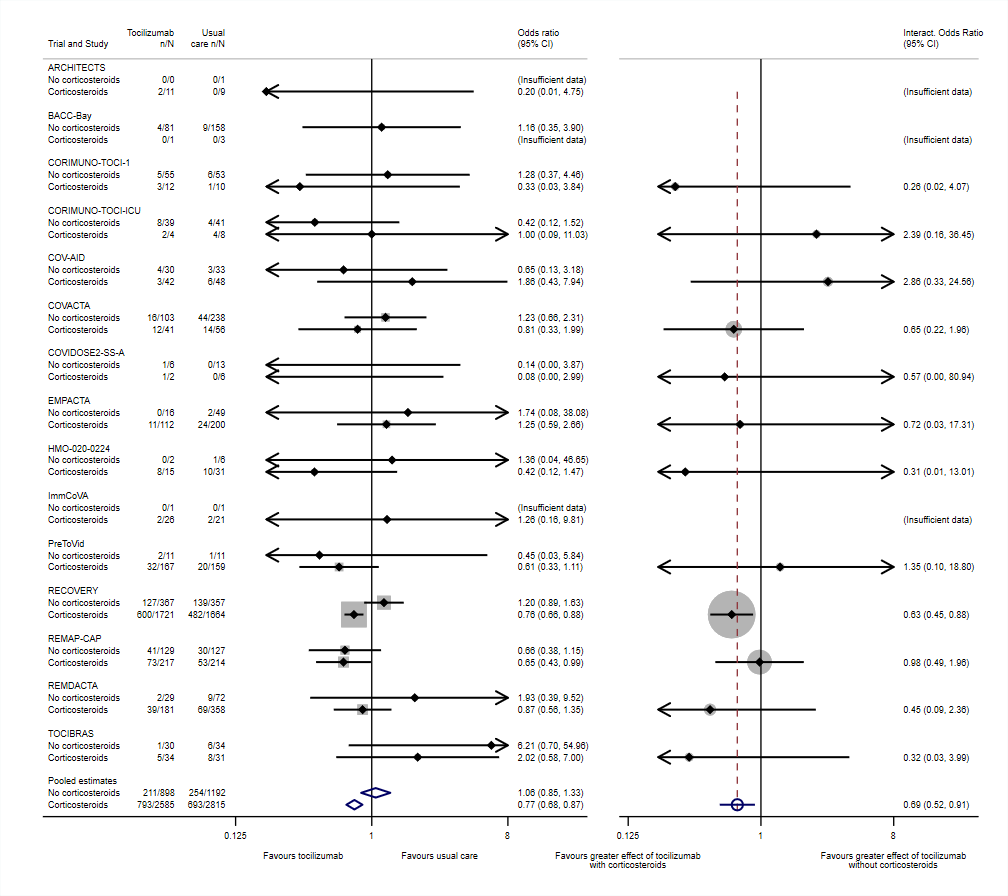
A "two-panel" subgroup and interaction forest plot, as proposed by Godolphin et al. (2023)
Data taken from Supplementary Appendix 2, and displayed in Figure 1, of Godolphin et al (as above). Data originally presented in Supplement 2, page 14, of Shankar-Hari et al. (2021).
Note that this same example is used to demonstrate the metan and forestplot commands within the metan GitHub pages, but without using metafloat directly (and without generating subgroup-specific treatment effects free from aggregation bias). It may be seen that far more lines of code are needed.
To run this example in full, the forestplot command is required. This command is part of the metan package, which may be installed by typing:
. net describe metan, from("https://raw.githubusercontent.com/UCL/metan/master/src/")
* Example generated by -dataex-. For more info, type help dataex
clear
input str17 TrialName byte Subgroup int(n0 e0 n1 e1)
"ARCHITECTS" 0 0 0 1 0
"ARCHITECTS" 1 11 2 9 0
"BACC-Bay" 0 81 4 158 9
"BACC-Bay" 1 1 0 3 0
"CORIMUNO-TOCI-1" 0 55 5 53 6
"CORIMUNO-TOCI-1" 1 12 3 10 1
"CORIMUNO-TOCI-ICU" 0 39 8 41 4
"CORIMUNO-TOCI-ICU" 1 4 2 8 4
"COV-AID" 0 30 4 33 3
"COV-AID" 1 42 3 48 6
"COVACTA" 0 103 16 238 44
"COVACTA" 1 41 12 56 14
"COVIDOSE2-SS-A" 0 6 1 13 0
"COVIDOSE2-SS-A" 1 2 1 6 0
"EMPACTA" 0 16 0 49 2
"EMPACTA" 1 112 11 200 24
"HMO-020-0224" 0 2 0 6 1
"HMO-020-0224" 1 15 8 31 10
"ImmCoVA" 0 1 0 1 0
"ImmCoVA" 1 26 2 21 2
"PreToVid" 0 11 2 11 1
"PreToVid" 1 167 32 159 20
"RECOVERY" 0 367 127 357 139
"RECOVERY" 1 1721 600 1664 482
"REMAP-CAP" 0 129 41 127 30
"REMAP-CAP" 1 217 73 214 53
"REMDACTA" 0 29 2 72 9
"REMDACTA" 1 181 39 358 69
"TOCIBRAS" 0 30 1 34 6
"TOCIBRAS" 1 34 5 31 8
end
generate long f1 = n1 - e1
generate long f0 = n0 - e0
quietly metan e1 f1 e0 f0, or nogr keeporder
label variable Trial Trial
label variable Subgroup `""Corticosteroid use""'
metafloat _ES _seES, study(Trial) subgroup(Subgroup) fixed clear keepvars(e1 n1 e0 n0)
gen counts0 = strofreal(cond(missing(e0), 0, e0)) + "/" + strofreal(cond(missing(n0), 0, n0)) if inlist(_y_USE, 1, 2)
gen counts1 = strofreal(cond(missing(e1), 0, e1)) + "/" + strofreal(cond(missing(n1), 0, n1)) if inlist(_y_USE, 1, 2)
label variable counts0 `""Usual care" "n/N""'
label variable counts1 `""Tocilizumab" "n/N""'
forestplot, prefix(_y) labels(_LABELS) or nowt lcols(counts0 counts1) keepall ///
favours("Favours tocilizumab" " " # "Favours usual care" " ") ///
xlabel(.125 "0.125" 1 8) range(.125 8) cirange(.2 8) boxsca(50) texts(150) astext(50) savedims(A) graphregion(color(white)) scheme(s2color) name(left)
forestplot, prefix(_yInt) interaction usedims(A) nonames eform effect("Interact. Odds Ratio") keepall nowt ///
favours("Favours greater effect of tocilizumab" "with corticosteroids" ///
# "Favours greater effect of tocilizumab" "without corticosteroids", fp(8)) ///
xlabel(.125 "0.125" 1 8) range(.125 8) cirange(.2 8) texts(150) graphregion(color(white)) scheme(s2color) name(right)
graph combine left right, imargin(zero) xsize(4.5) graphregion(color(white)) scheme(s2color)