Given the trajectories of neuronal fiber pathways, we model the evolution of trajectories that capture geometrically significant events (akin to a
fingerprint) and calculate their point correspondence in the 3D brain space. We show that our model can handle the presence of improbable streamlines, as commonly produced by tractography algorithms. Three key parameters in our method capture the geometry and topology of the streamlines: (i)
For more details about our algorithm, please refer to our paper.
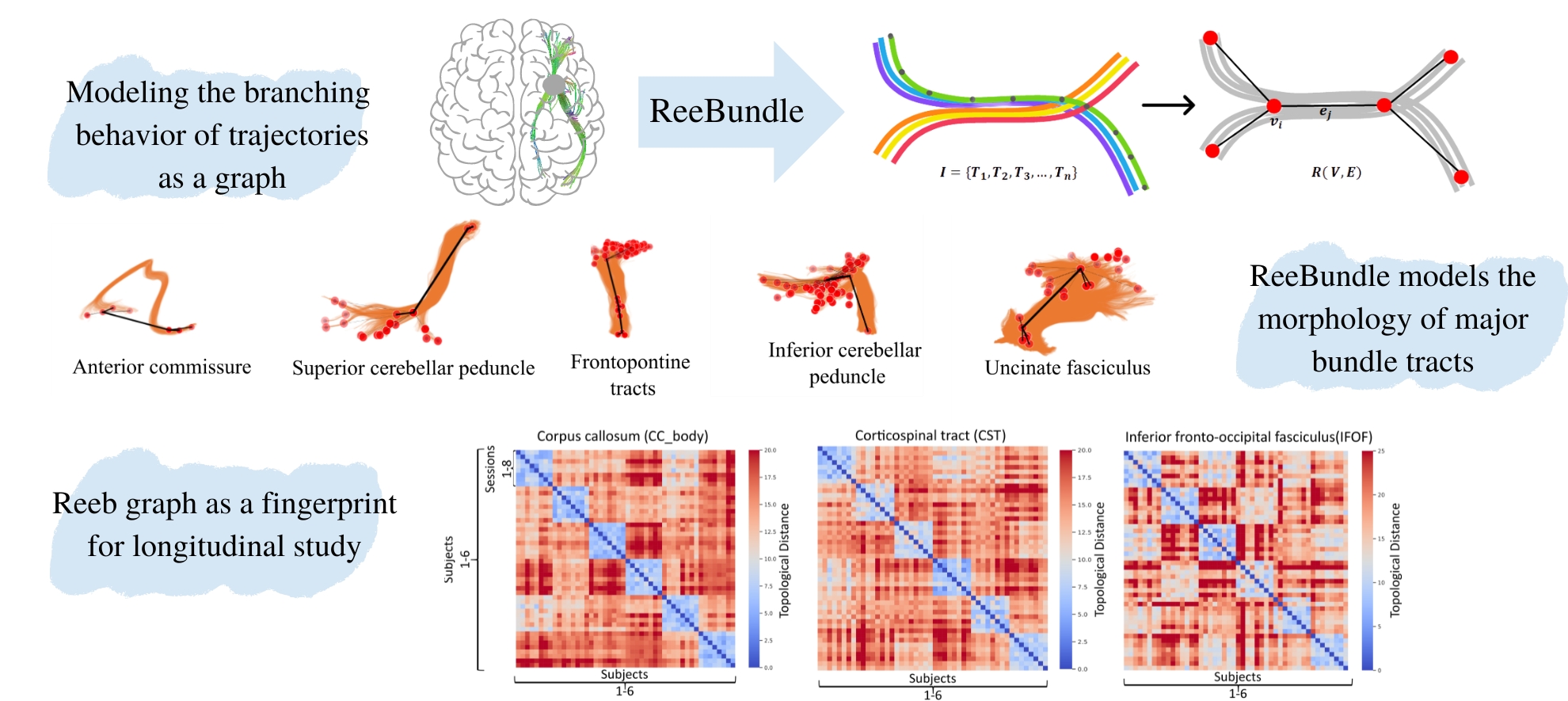
The system was employed for our research presented in [1], where we propose a novel and efficient algorithm to model high-level topological structures of neuronal fibers. Tractography constructs complex neuronal fibers in three dimensions that exhibit the geometry of white matter pathways in the brain. However, most tractography analysis methods are time consuming and intractable. We develop a computational geometry-based tractography representation that aims to simplify the connectivity of white matter fibers. If the use of the software or the idea of the paper positively influences your endeavours, please cite [1] and [2].
[1] Shailja, S., Angela Zhang, and B. S. Manjunath. "A computational geometry approach for modeling neuronal fiber pathways." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2021.
[2] Shailja, S., Bhagavatula, V., Cieslak, M., Vettel, J., Grafton, S. T., and B. S. Manjunath. "ReeBundle: a method for topological modeling of white matter pathways. " Submitted to the Journal, 2022.
To download all prerequisites, in the terminal type
pip install -r requirements.txt
The code has been tested only on python version 3.7.
Visualize an example Reeb graph in 3D.
-
For ISMRM Dataset, sample .trk files are included in teh SampleISMRMData folder. Run the jupyter notebook "ReebGraphForISMRMBundles" to load the .trk files and construct the Reeb graphs for CA and CP bundle tracts. For ISMRM, ReeBundle detects and handles the morphology of the white matter tract configurations due to branching and local ambiguities in complicated bundle tracts like anterior and posterior commissures. ISMRM dataset helps in the qualitative validation of the ReeBundle method.
-
For the CRASH Dataset, sample .gpickle files for Reeb graphs can be found in SampleISMRMData. Run the jupyter notebook "TopologicalDistanceForCRASH" to load .gpickle files and compute topological distances. Finally, creates the heatmap for CC bundle as presented in the paper [2]. In the analysis of the bundle tracts of white matter fibers in the CRASH dataset, the Reeb graphs of a given subject are similar across eight scan repetitions acquired weeks apart, whereas they differ from other subjects. CRASH dataset helps in the quantitative validation of the Reeb graph as a fingerprint for the bundle tract.
-
For Tractometry methods, values of the features computed for CC ROIs can be found in the 'CCtractometry.csv'. Run the jupyter notebook "" to load the tractometry file and compute the inter- and intra- distances. The statistical significance of the difference between \emph{inter-} and \emph{intra-} distances based on these tractometry metrics is negligible
$(p \sim .99)$ whereas ReeBundle provides a graph representation that visualizes, quantifies, and localizes the topological differences.
The code was tested on the publicly available ISMRM dataset and the proposed Reeb graph-based distance was tested on CRASH dataset.