Automatic mapping of German medical-related terms into SNOMED-CT.
Currently, this repository is under development for code quality improvement.
This repository contains the source code used in the development of our mapping approach between German medical phrases and The Observational Medical Outcomes Partnership (OMOP) concepts.
Compared to the current standard (USAGI) our approach performs slightly better. Its main advantage lies in the automatic processing of German phrases to English OMOP concept suggestions, operating without the need for human intervention.
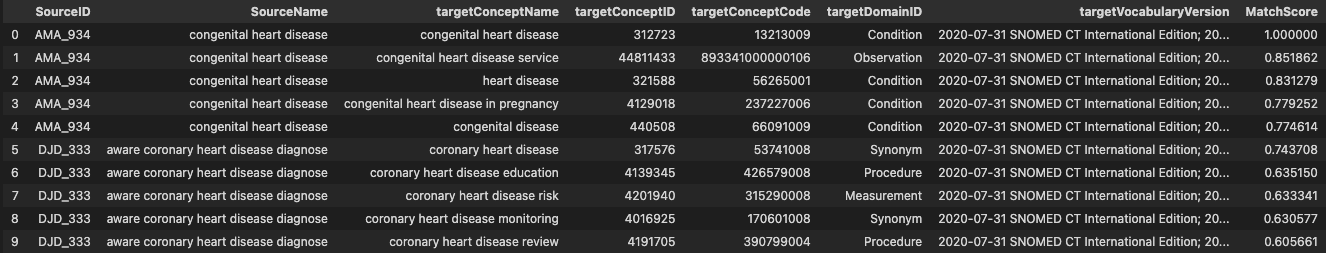
Example of the result after translation and mapping (column names in USAGI format)
This project utilizes Docker for providing an OS-independent development and integration experience. We highly recommend using Visual Studio Code and the associated "Development Container" which allows direct access to an environment and shell with pre-installed Python, corresponding packages, and a specialized IDE experience. However, running Docker standalone is also possible by using the Dockerfile in the devcontainer folder.
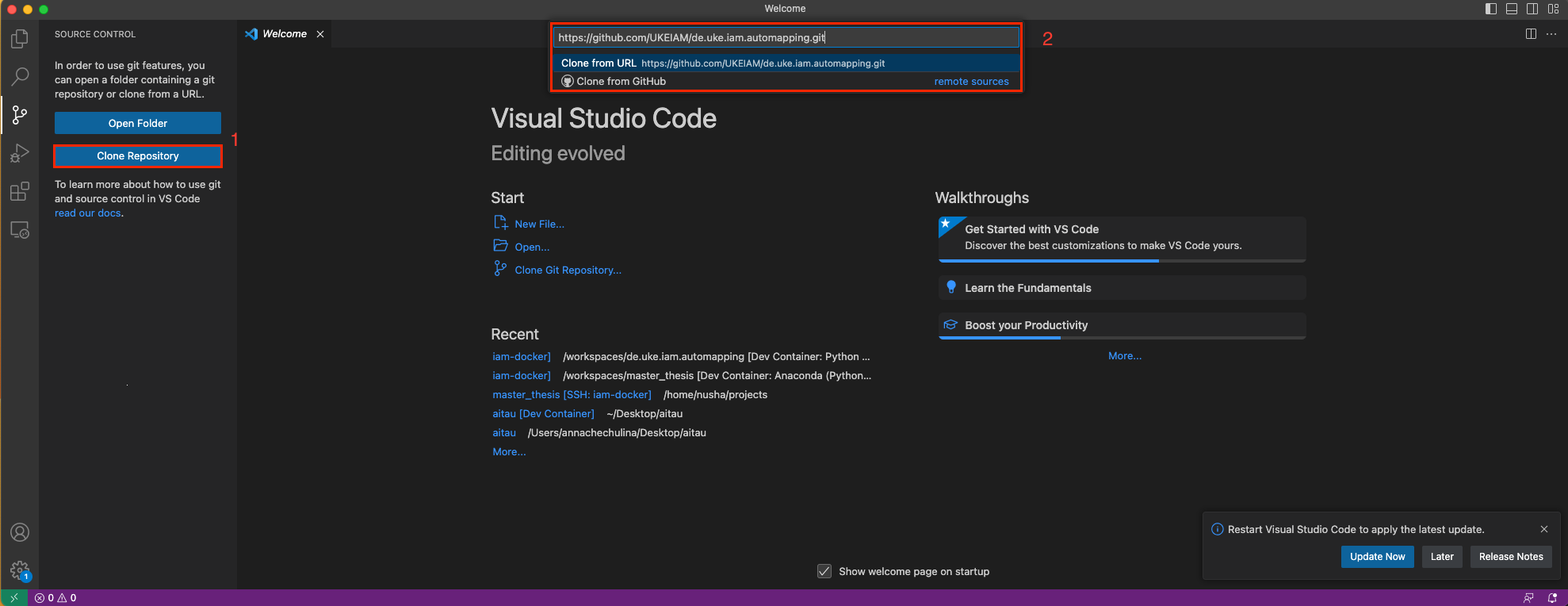
- Clone repository (we recommend using Visual Studio Clone)
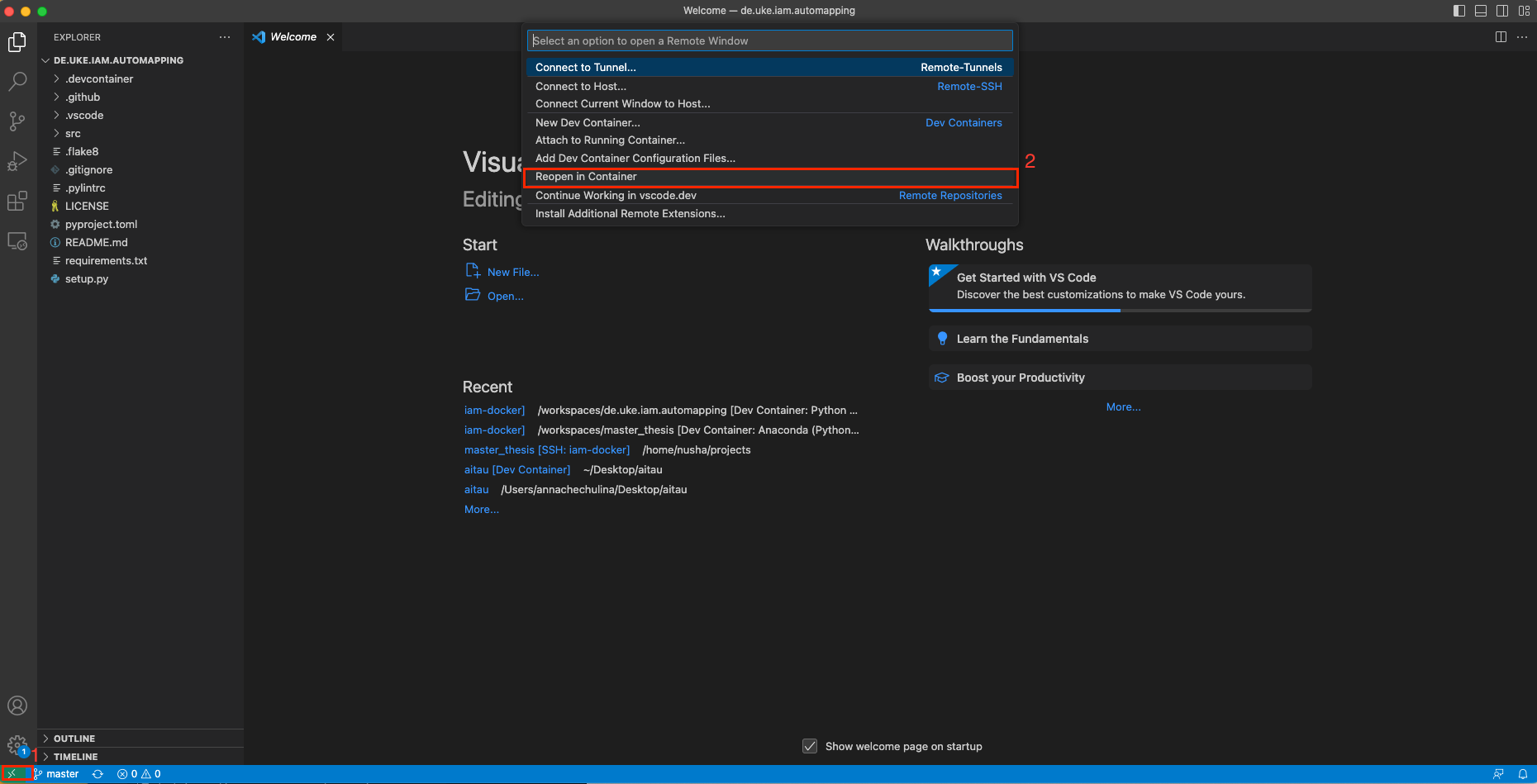
- Reopen the Repository in the Docker container
-
Download OMOP CDM files from [Athena] (https://athena.ohdsi.org/search-terms/start) with SNOMED CT vocabulary
-
From Downloaded Archiv put files: CONCEPT.csv, CONCEPT_SYNONYM.csv, and VOCABULARY.csv in a folder with the repository
-
Open example pipeline and replace variables with yours (path to files)
| Variable Name | Description |
|---|---|
| data_file | Path to Excel file with your data(should contain columns with id and name) |
| abbreviation_file | Path to Excel file with german medical abbreviation and descriptions |
| concept_file | Path to the file CONCEPT.csv from Athena |
| synonym_file | Path to the file CONCEPT_SYNONYM.csv from Athena |
| vocabulary_file | Path to the file VOCABULARY.csv from Athena |
- Run updated pipeline
|- data_example/ (folder with data used in the pipeline)
|- example_data.xlsx (example of data used to run the pipeline)
|- german_abbreviation.xlsx (table with German medical abbreviations and their description)
|- mapping.csv (a result from the pipeline)
|- notebooks/
|- pipeline.ipynb (Jupyter Notebook with an example of how to run source code)
|- src
|- automapping/ (source code)
|- concept.py (object to keep concept)
|- concepts.py
|- detection.py (object to create a data frame with predictions)
|- language.py
|- m5.py (integration with internal application)
|- loader.py (file with data loading)
|- prediction.py (object with keep predictions)
|- preprocessor.py (abbreviation replacement and nlp preprocessing)
|- translator.py (translation with Hugging Face model from German to English)
|- mapper.py (object with TF-idf model)
|- sample.py (object with a sample of the given data)
config.yaml (config for flask application)
flask_application_endpoint_swagger.py (integration of this pipeline with internal application)
This project is licensed under the GPL-3.0 license. Everyone is permitted to copy and distribute verbatim copies of this license document, but changing it is not allowed.
See LICENSE for more information.