The NExtSteP (NRL Extensible Stealthy Protocol) Testbed enables the experimental analysis of the detectability of potentially stealthy communications. The testbed ("NExtSteP" for short) is extensible in that it allows various types of innocuous cover traffic, various methods of embedding communication into that traffic, various methods to attempt to detect embedded communication, and various metrics for evaluating the performance of these detectors.
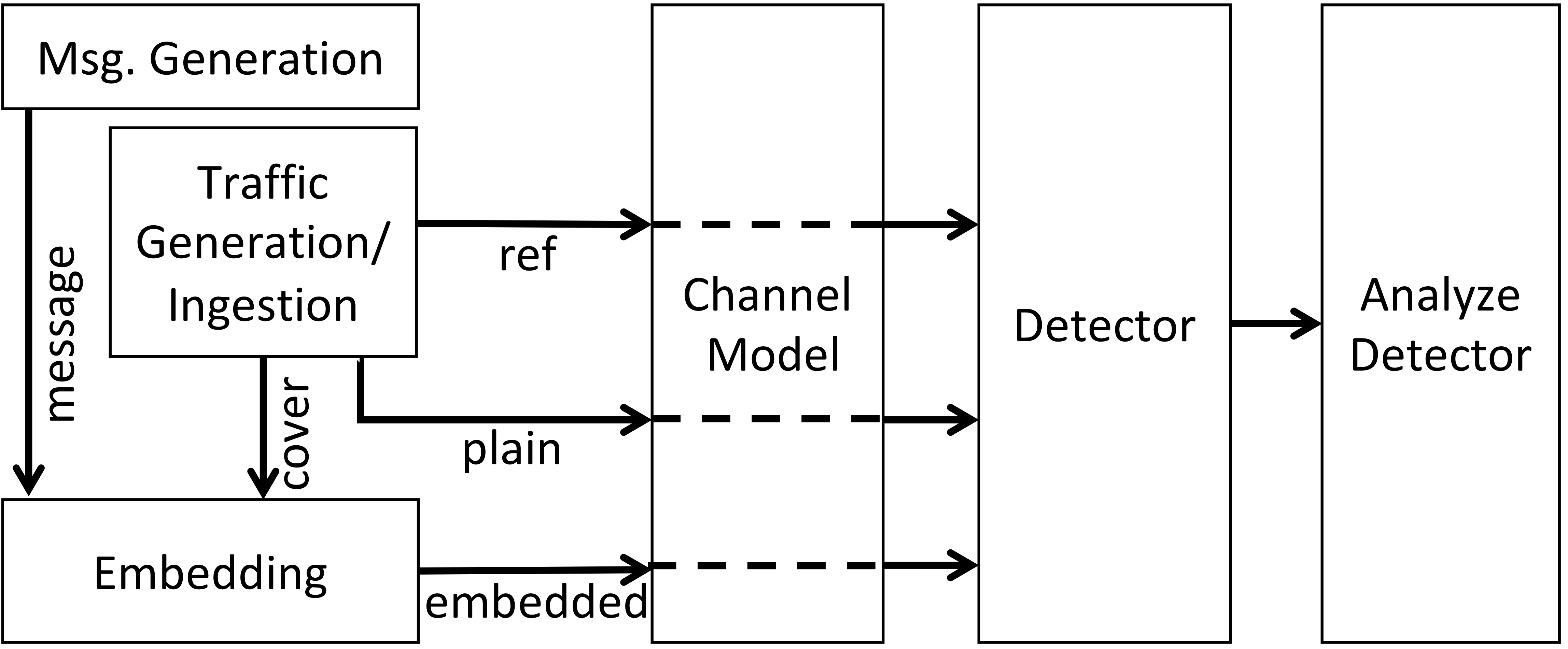
The output of a NExtSteP experiment is an evaluation of a detector against an embedding method (in a particular traffic setting). The process for getting to that point is illustrated in the following figure.
In particular, NExtSteP produces a message, modeled as a random bit sequence (to mimic an encrypted message). It also produces three types of traffic samples: "reference", "cover", and "plain". These can be generated synthetically by NExtSteP, or they can be ingested by NExtSteP from external captures. NExtSteP then embeds some part of the message into the cover traffic, using a specified embedding method, to produce "embedded" traffic. The reference, plain, and embedded traffic is passed through a channel model; this may leave it unmodified, add synthetic noise (a future step), or add noise via emulation of a network (currently envisioned to be done using CORE). The traffic is then provided to a detector, which must distinguish plain from embedded traffic; typically, detectors make use of reference traffic to do this. Finally, the results of the detector (over many different examples of plain and embedded traffic, and potentially with respect to multiple examples of reference traffic) are evaluated using various metrics.
If the NExtSteP repository (which we will call the "nextstep directory") is nextstep/, create the following directories manually in the same directory as nextstep/:
data/
data/logs/
data/sqrtdata/
This version of NExtSteP has been developed primarily under Python 3.7.1.
The packages used are described in the pip requirements file nextstep/requirements.txt.
NExtSteP allows the use of various PRNGs. The configuration file nextstep/nextstep.cfg in this release uses ChaCha20 from the PyCryptodome package (included in the pip requirements file). The code may also assume the presence of the following PRNGs (these should be installed manually or references to them may need to be removed from the NExtSteP code):
PCG (minimal C implementation, version 0.9), available from http://www.pcg-random.org/ The distributed pcg-c-basic-0.9/ directory should be placed in nextstep/libStealthy/pRandom/
SFMT (v. 1.5.1), available from http://www.math.sci.hiroshima-u.ac.jp/~m-mat/MT/SFMT/index.html The distributed SFMT-src-1.5.1/ directory should be placed in nextstep/libStealthy/pRandom/
NExtSteP includes a slightly restructured version of the public-domain xoshiro256 code (original available from http://prng.di.unimi.it/).
After doing the setup described above, run
python buildExt.py
in the nextstep/ directory to complete the installation.
NExtSteP scripts can be largely controlled through the command line as illustrated in the sample shell scripts discussed below. Options that are not given on the command line are typically controlled by a configuration file whose location may be specified on the command line. A default version of that file is at nextstep/nextstep.cfg. Options specified on the command line take precedence over those specified int the configuration file; within the configuration file, options specified in a script-specific section take precendence over those specified in the [general] section. A handful of parameters (including the location of the configuration file) have default values that are used if they are unspecified on the command line.
The script nextstep/tools/generate-msgs.py produces a file with random bits for use as a message to embed. (The randomness mimics an encrypted message.) This produces a file nextstep/urand-b<N>.txt containing <N> random bits using Python's os.urandom function. The default is to produce 4,000,000 bits; for other sizes, the code needs to be edited to change the argument to the genRandom function.
The included data file nextstep/urand-b4000000.txt can be used instead of running this script.
The script nextstep/synthetic/generate-data.py is used to generate synthetic IPD sequences drawn from a specified distribution. Running it with the --refs flag produces reference sequences; running it without this flag produces both plain and cover sequences. All sequences are drawn from the same distribution, but the file names and locations differ.
Options include (but are not limited to) the distribution from which IPD values should be chosen and the number of sequences to produce. See the script help (invoked by python generate-data.py --help) for further information.
Alternatvely to synthetic generation, the script nextstep/ingest/ingest-data.py is used to produce reference, plain, and cover IPD sequences from ingested data. The data to ingest can be given as an argument to the script. It should be a JSON-encoded list of IPD values.
The script nextstep/experiments/embed.py is used to embed the (some portion of the) generated message into the cover sequences in order to produce embedded sequences. The options for this include (but are not limited to) parameters to indicate which cover sequences to use, the embedding method to use, and various embedding parameters. See the script help (invoked by python embed.py --help) for further information.
The script nextstep/experiments/classify.py is used to run detectors. The output of a classifier is a sequence of tuples; each tuple contains statistics about an individual plain or embedded sequence and an indication of whether the sequence contained an embedded message. For detectors that make use of a reference sequence, there is one tuple sequence per reference sequence. Results are stored in data/sqrtdata/hq/classout/ (results from synthetic data) or data/sqrtdata/ingest/classout/ (results from ingested data).
The options for this include (but are not limited to) parameters to indicate which plain/embedded sequences to use, the detector to use, and any detector parameters. See the script help (invoked by python classify.py --help) for further information.
The script nextstep/experiments/analyze.py is used to evaluate detectors based on the classification output. It produces a plot of the performance of the detector as a function of the length of the plain/embedded sequences. These are stored in data/sqrtdata/hq/analyzeout/ (plots for synthetic data) or data/sqrtdata/ingest/analyzeout/ (plots for ingested data).
The options for this include (but are not limited to) options for the type of evaluation (the metric), plotting options, and the data that should be plotted. See the script help (invoked by python analyze.py --help) for further information.
This repository includes sample shell scripts and data to run NExtSteP.
The sample shell scripts call the individual Python scripts described above. The nextstep/urand-b4000000.txt file contains 4,000,000 bits output by generate-msgs.py, so that Python script is not called by the sample shell scripts.
The nextstep/synthetic/run-refs-SAMPLE.sh shell script generates synthetic reference, cover, and plain data.
The nextstep/experiments/run-exp-SAMPLE.sh shell script does embedding, classification using detectors, and detector evaluation for synthetic data. It omits modification using a noisy channel model; this may be thought of as using a noiseless channel.
The nextstep/ingest/run-ingest-SAMPLE.sh shell script ingests data (from a location specified near the top of the script) and then runs the embedding, classification, and analysis scripts. This covers the entire process for ingested data. As with the synthetic example, this omits the step of modeling a noisy channel.
NExtSteP is experimental and still very much under development. Anything---file formats/names/locations, algorithms, testbed structure, etc.---may be changed in future releases.
See nextstep/LICENSE.TXT for license information.
The NExtSteP testbed is designed and written by Olga Chen, Aaron D. Jaggard, Catherine Meadows, and Michael Shlanta (U.S. Naval Research Laboratory). We thank David Li (MITRE) and Sarah Mousley Mackay (LLNL) for additional contributions they made while at NRL.
The NExtSteP team may be reached via nextstep@nrl.navy.mil