Development version
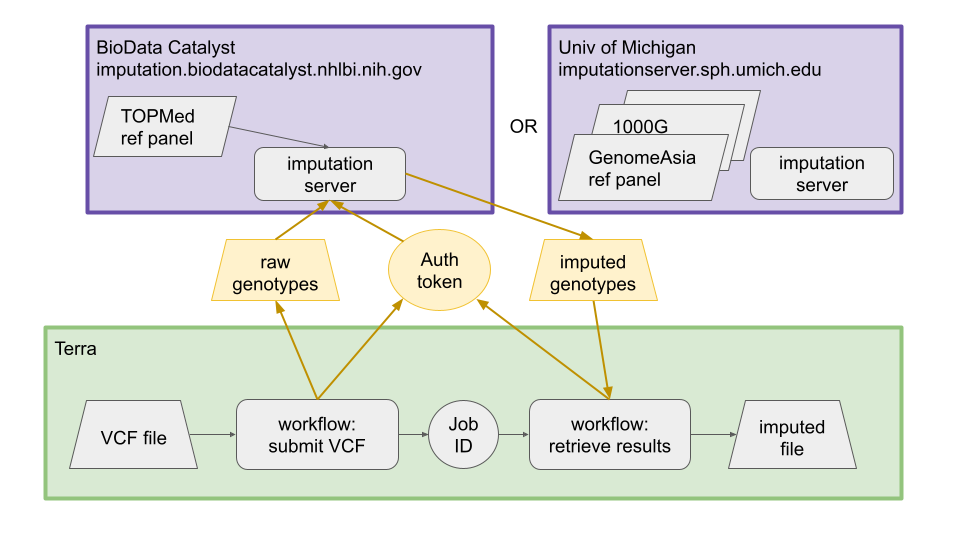
This repository is for WDL workflows to submit jobs to the NHLBI BioData Catalyst TOPMed and Michigan imputation servers. This is a work in progress and not yet intended for general use.
The Michigan imputation server and the TOPMed imputation server are cloud instances of the same imputation software with different reference panels. The imputationbot software allows a user to submit files to either server using the command line. Users must create an account on the server they wish to use, then download an authentication token. VCF files are uploaded to the server, imputed, and the results are available for download for 7 days.
The Dockerfile creates a docker image containing the imputationbot software. The image is available on Docker Hub as uwgac/primed-imputation.
The workflows are written in the Workflow Description Language (WDL). This GitHub repository contains the Dockerfile, the WDL code, and JSON files containing inputs to each workflow, both for testing and to serve as examples. The bash scripts were used for testing imputationbot and are not used in the workflows.
imputationbot has a command to register a token interactively; however, this command writes the token to a file in the user’s home directory. The WDLs take the token as an input string and write the configuration file directly, before running any other commands.
The user must specify the following inputs:
| input | description |
|---|---|
| token | string with the authentication token (note the example in the JSON file is not a real token) |
| hostname | URL for either the TOPMed or Michigan server |
| refpanel | “topmed-r3” is the only option for TOPMed, but there are multiple options for Michigan (see below) |
| population | “all” for TOPMed, multiple options for Michigan (see below) |
| vcf_files | files to impute. if multi_chrom_file is true, only one file should be provided. |
| multi_chrom_file | boolean, set to true if vcf_files contains a single file with multiple chromosomes; false if vcf_files are already split by chromosome |
| build | genome build of the input files, hg19 or hg38 |
| r2_filter | r2 filter to be applied to the results. Default is 0, other possible values are 0.001, 0.1, 0.2, 0.3 |
| meta_imputation | boolean for whether to generate a meta-imputation file. Default is true. |
| password | string that must also be supplied to the results workflow for download. Specifying the password during job submission means the user doesn’t have to rely on receiving the password by email. |
When VCF files are submitted to the imputation server, a job_id is assigned. The submit workflow returns this job_id as an output, and it must be provided to the results workflow.
The workspace specified by workspace_name and workspace_namespace must already contain the subject, sample, and sample_set tables. The resulting files are added as imputation_dataset and imputation_file tables.
The user must specify the following inputs:
| input | description |
|---|---|
| token | string with the authentication token (note the example in the JSON file is not a real token) |
| hostname | URL for either the TOPMed or Michigan server |
| job_id | string returned by the submission workflow |
| password | string that must also be supplied to the results workflow for download. Specifying the password during job submission means the user doesn’t have to rely on receiving the password by email. |
| disk_gb | Disk size (in GB) required. If in doubt, consult the Jobs page of the imputation server to view the total file size of the results. |
| refpanel | The reference panel used for imputation (same value as from imputation_server_submit) |
| r2_filter | r2 filter that was applied to the results (same value as from imputation_server_submit) |
| sample_set_id | The sample_set_id of the dataset that was imputed |
| source_dataset_id | The array_dataset_id of the dataset that was imputed |
| source_genotypes | A description of the array used for the dataset that was imputed |
| model_url | A URL providing the path to the data model in JSON format. |
| import_tables | A boolean indicating whether data model tables should be imported to the workspace. |
| overwrite | A boolean indicating whether existing rows in the workspace data tables should be overwritten. |
| workspace_name | A string with the workspace name. e.g, if the workspace URL is https://anvil.terra.bio/#workspaces/fc-product-demo/Terra-Workflows-Quickstart, the workspace name is "Terra-Workflows-Quickstart" |
| workspace_namespace | A string with the workspace name. e.g, if the workspace URL is https://anvil.terra.bio/#workspaces/fc-product-demo/Terra-Workflows-Quickstart, the workspace namespace is "fc-product-demo" |
| vcf_disk_gb | Disk space required for each VCF file (default 10 GB). If the job fails due to lack of disk space, try setting this to a larger value. |
The imputed genotypes and accompanying files (log, QC report, statistics, md5) are downloaded to the user’s workspace.
?????????????????????????????????????????????????????????????????????????????????
? ID ? Name ? Populations ? Instance ?
?????????????????????????????????????????????????????????????????????????????????
? topmed-r3 ? TOPMed r3 ? all vs. TOPMed Panel ? TOPMed Imputation Server ?
? ? ? mixed Skip ? ?
?????????????????????????????????????????????????????????????????????????????????
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? ID ? Name ? Populations ? Instance ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? 1000g-phase-1 ? 1000G Phase 1 v3 Shapeit2 (no singletons) (GRCh37/hg19) ? afr AFR ? Michigan Imputation Server ?
? ? ? amr AMR ? ?
? ? ? asn ASN ? ?
? ? ? eur EUR ? ?
? ? ? mixed Other/Mixed ? ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? 1000g-phase3-low ? 1000G Phase 3 GRCh38 (BETA) ? all ALL ? Michigan Imputation Server ?
? ? ? mixed Other/Mixed ? ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? 1000g-phase3-deep ? 1000G Phase 3 GRCh38 30x (BETA) ? all ALL ? Michigan Imputation Server ?
? ? ? mixed Other/Mixed ? ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? 1000g-phase-3-v5 ? 1000G Phase 3 v5 (GRCh37/hg19) ? afr AFR ? Michigan Imputation Server ?
? ? ? amr AMR ? ?
? ? ? eas EAS ? ?
? ? ? sas SAS ? ?
? ? ? eur EUR ? ?
? ? ? mixed Other/Mixed ? ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? caapa ? CAAPA African American Panel (GRCh37/hg19) ? AA African Americans ? Michigan Imputation Server ?
? ? ? mixed Other/Mixed ? ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? genome-asia-panel ? Genome Asia Pilot - GAsP (GRCh37/hg19) ? asn ASN ? Michigan Imputation Server ?
? ? ? mixed Other/Mixed ? ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? hapmap-2 ? HapMap 2 (GRCh37/hg19) ? eur EUR ? Michigan Imputation Server ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????
? hrc-r1.1 ? HRC r1.1 2016 (GRCh37/hg19) ? eur EUR ? Michigan Imputation Server ?
? ? ? mixed Other/Mixed ? ?
??????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????????