BrAPI Graphical Filtering 

- Try it out here
- Include-ready version can be found under releases.
- Also available via npm:
npm install @solgenomics/brapi-graphical-filtering
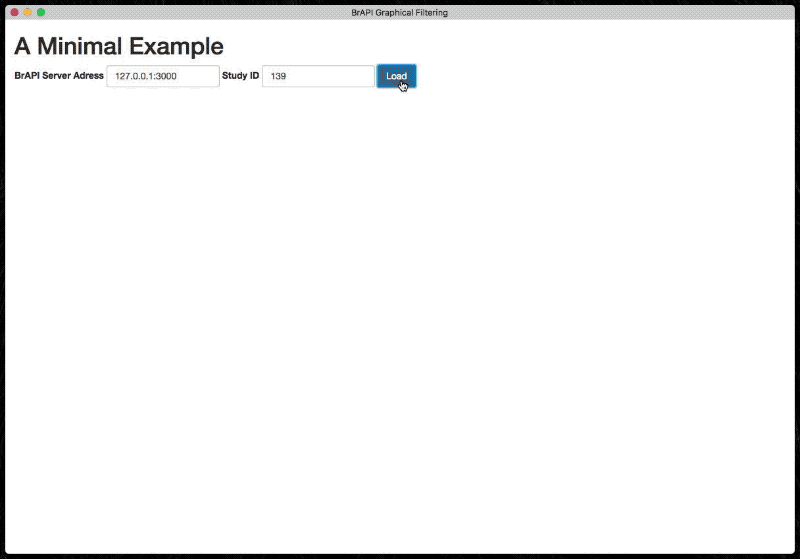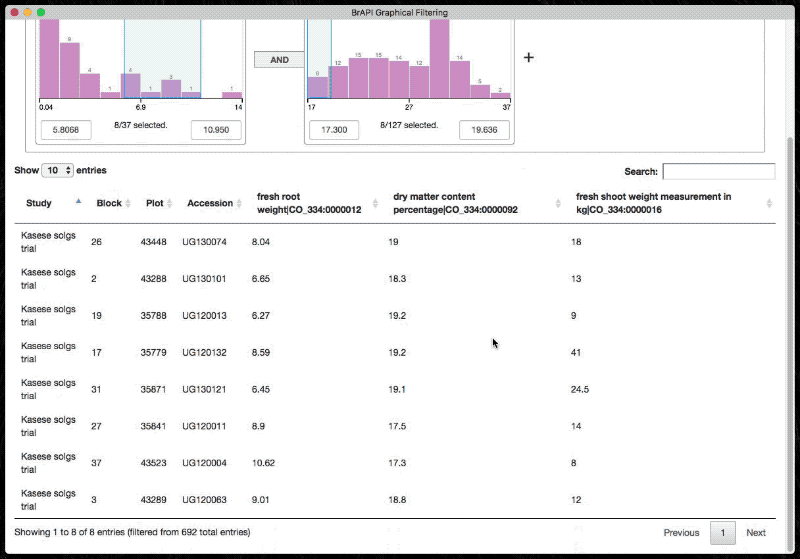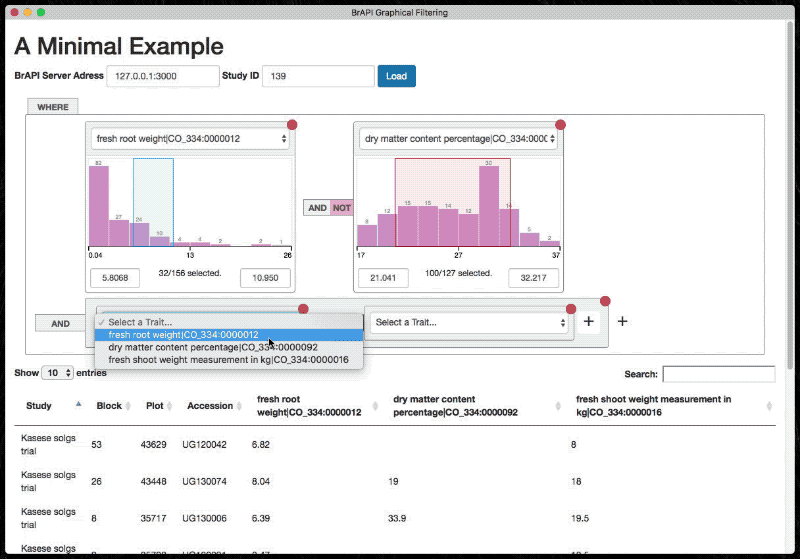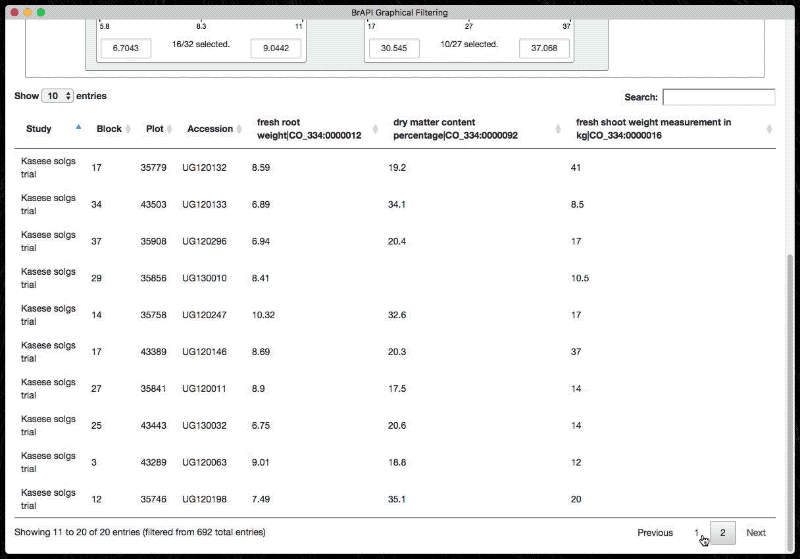
The graphical filter exposes a minimal API for initializing and populating the table. It is designed to rely on BrAPI.
Use the following snippets (in order) to fully set up a tree.
- Create a new GraphicalFilter object
var gf = GraphicalFilter( // BrAPI search of observationUnits to be displayed BrAPI("https://brapi.myserver.org/brapi/v1").phenotypes_search(myparams), // Accessor describing traits for each observationUnit (returns object) function(d) { var traits = {} d.observations.forEach(function(obs){ traits[obs.observationVariableName] = obs.value; }); return traits; }, // Accessor describing extra columns to display in the table (returns object) function(d) { return { 'Accession':d.germplasmName } }, // Order to display the above columns in (array) ["Accession"], // key to group observationUnits by in the table (key value or undefined for no grouping) function(d) { // groupBy function return d.germplasmDbId } );
- Then, draw the filter and table.
gf.draw( // div to draw the filter in "#filter_div", // table to output filtered data to "#filtered_results" );
- Bootstrap (3.3) (CSS & JS)
- D3.js (4)
- DataTables (1) (JS)
- jQuery (3)
- @solgenomics/brapp-wrapper (For building the example.)