This is the repository for the code and raw data of the paper entitled:
Modelling intercropping functioning to support sustainable agriculture
If you're interested in reproducing the simulations, prefer downloading the full repository from Zenodo that provides our version of JavaStics (it is not given here to save space).
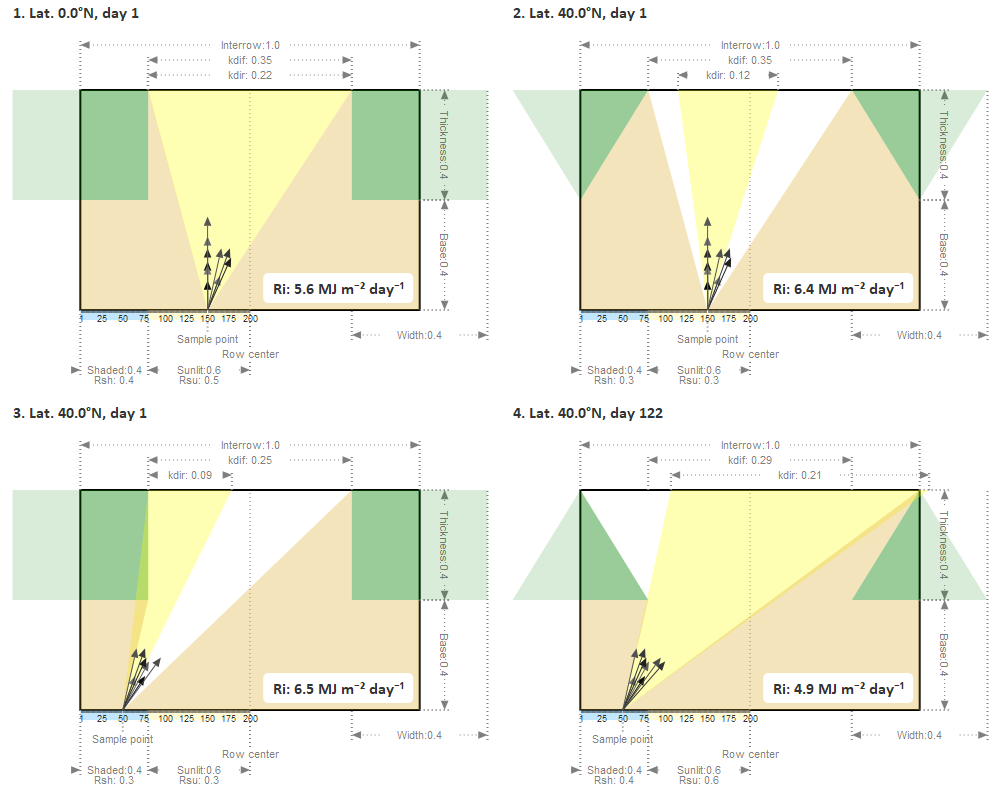
This repository contains the Pluto notebook that you can use to better understand how the radiative transfer computation works, which was used to produce Figure A1 from the article (see below). To use it, please head over this link: https://vezy.github.io/STICS-IC-paper. Then you can run it on binder (it can be very slow to open the first time), or download it and run it using Julia (you can download Julia here):
# add Pluto # uncomment this line if you run it for the first time, it will download and install the Pluto package
using Pluto
Pluto.run(notebook="index.jl")Figure 1. Diagram of the light interception computation using the radiative transfer option. See scientific article for more information.
You can remake all the steps from the article using the code and data in this repository. Be careful though, the optimization steps can take quite some time to execute so it may be best to use a cluster to run the simulations.
The optimization results are available from the usms-optim-beer and usms-optim-radiative repository for the optimization using the Beer-Lambert law of light extinction and the radiative transfer options respectively.
Here are some explanations about the different folders you can find in this project and what are they used for:
├───.github
│ └───workflows # Used to build automatically the notebook and make a website out of it.
├───0-data
│ ├───citations # Compute STICS citations (but I used a different, curated database from the STICS team instead)
│ ├───usms # The simulations we use, curated and checked, with original parameter values from the articles or the general database
│ │ ├───1Tprecoce2Stardif2012 # USM for the intercrop of sunflower and soybean in 2012, No N
│ │ ├───Angers-IC-Pea_Barley # USM for the intercrop of pea and barley in Angers, No N
│ │ ├───Angers-SC-Barley # USM for the sole crop of barley in Angers, No N
│ │ ├───Angers-SC-Pea # USM for the sole crop of pea in Angers, No N
│ │ ├───Auzeville-IC # USM for the intercrop of pea and wheat in Auzeville (alternate row) in 2005-2006, No N
│ │ ├───Auzeville-IC-2012 # USM for the intercrop of pea and wheat in Auzeville (mixed in the row) in 2012-2013, with 140kg mineral N
│ │ ├───Auzeville-Pea-2012-SC # USM for the sole crop of pea in Auzeville in 2012-2013, with 140kg mineral N
│ │ ├───Auzeville-Pea-SC # USM for the sole crop of pea in Auzeville in 2005-2006, no N
│ │ ├───Auzeville-Wheat-2012-SC # USM for the sole crop of wheat in Auzeville in 2012-2013, with 140kg mineral N
│ │ ├───Auzeville-Wheat-SC # USM for the sole crop of wheat in Auzeville in 2005-2006, no N
│ │ ├───Auzeville_wfb-Fababean-SC # USM for the sole crop of fababean in Auzeville in 2006-2007, no N
│ │ ├───Auzeville_wfb-Fababean-Wheat-IC # USM for the intercrop of fababean and wheat in Auzeville (alt. row) in 2006-2007, no N
│ │ ├───Auzeville_wfb-Wheat-SC # USM for the sole crop of wheat in Auzeville in 2006-2007, no N
│ │ ├───sojaTardif2012-SC # USM for the sole crop of soybean in Auzeville in 2012-2013, no N
│ │ └───tourPrecoce2012-SC # USM for the sole crop of sunflower in Auzeville in 2012-2013, no N
│ ├───usms-optim-beer # = to "usms", but with plant parameters optimized using observations and the Beer-Lambert law of light extinction
│ └───usms-optim-radiative # Same folder than "usms", but with plant parameters optimized with the radiative transfer option
├───0-javastics # Full installation of JavaStics interface. Our version of the stics model is in the bin folder. You must download this folder from https://doi.org/10.5281/zenodo.6803363
├───0-stics # The code base for STICS-IC.
├───1-code # The code base for this project (mixed R and Julia source code). Files are numbered according to the execution step.
└───2-outputs # Outputs from the code in `1-code`.
Note: This repository works for Windows. If you are using Linux, you'll have to replace the windows version of STICS by the Linux version everywhere in the code base. Search "Stics_IC_v24-05-2022.exe" everywhere in the code and replace it by "Stics_IC_v24-05-2022_linux". Mac users should build their own executable from the 0-stics code base, and put it in the bin folder of JavaStics. you can find instruction for compilation here.