Gregory Holste, Ziyu Jiang, Ajay Jaiswal, Maria Hanna, Shlomo Minkowitz, Alan C. Legasto, Joanna G. Escalon, Sharon Steinberger, Mark Bittman, Thomas C. Shen, Ying Ding, Ronald M. Summers, George Shih, Yifan Peng, and Zhangyang Wang
MICCAI 2023, Early accept (top 14%!)
Pruning has emerged as a powerful technique for compressing deep neural networks, reducing memory usage and inference time without significantly affecting overall performance. However, the nuanced ways in which pruning impacts model behavior are not well understood, particularly for long-tailed, multi-label datasets commonly found in clinical settings. This knowledge gap could have dangerous implications when deploying a pruned model for diagnosis, where unexpected model behavior could impact patient well-being. To fill this gap, we perform the first analysis of pruning’s effect on neural networks trained to diagnose thorax diseases from chest X-rays (CXRs). On two large CXR datasets, we examine which diseases are most affected by pruning and characterize class “forgettability” based on disease frequency and co-occurrence behavior. Further, we identify individual CXRs where uncompressed and heavily pruned models disagree, known as pruning-identified exemplars (PIEs), and conduct a human reader study to evaluate their unifying qualities. We find that radiologists perceive PIEs as having more label noise, lower image quality, and higher diagnosis difficulty. This work represents a first step toward understanding the impact of pruning on model behavior in deep long-tailed, multi-label medical image classification. All code, model weights, and data access instructions can be found at https://github.com/VITA-Group/PruneCXR.
- NIH-CXR-LT:
- MIMIC-CXR-LT (requires credentialing through PhysioNet):
All trained models can be found at https://utexas.box.com/s/eenr2gumfjaazngvnwerhmo9c0qru23w under the trained_models directory. In this directory, you will find 60 subdirectories, representing the 30 "runs" of a ResNet50 trained on each dataset (NIH-CXR-LT and MIMIC-CXR-LT) as described in the paper. Each subdirectory contains the weights, training history, and evaluation metrics for a given run.
All test set predictions for each pruned model (used to generate all main figures and results in the paper) can be found at https://utexas.box.com/s/eenr2gumfjaazngvnwerhmo9c0qru23w under the nih-cxr-lt_L1-prune_preds and mimic-cxr-lt_L1-prune_preds directories. Each directory contains a .pkl file containing a NumPy array of test set predictions for each combination of seed
To reproduce the results in the paper:
- See Dataset Access to download NIH ChestXRay14 images with NIH-CXR-LT labels and MIMIC-CXR-JPG v2.0.0 images with MIMIC-CXR-LT labels. Place all 6 files in the
labelsdirectory of this repo. - Install prerequisite packages with Anaconda:
conda env create -f torch.ymlandconda activate torch. - (Optional) From https://utexas.box.com/s/eenr2gumfjaazngvnwerhmo9c0qru23w, download the
trained_modelsdirectory and place it in this repo's base directory. The script to train these models can be found insrc/run_experiments.sh. - From https://utexas.box.com/s/eenr2gumfjaazngvnwerhmo9c0qru23w, download
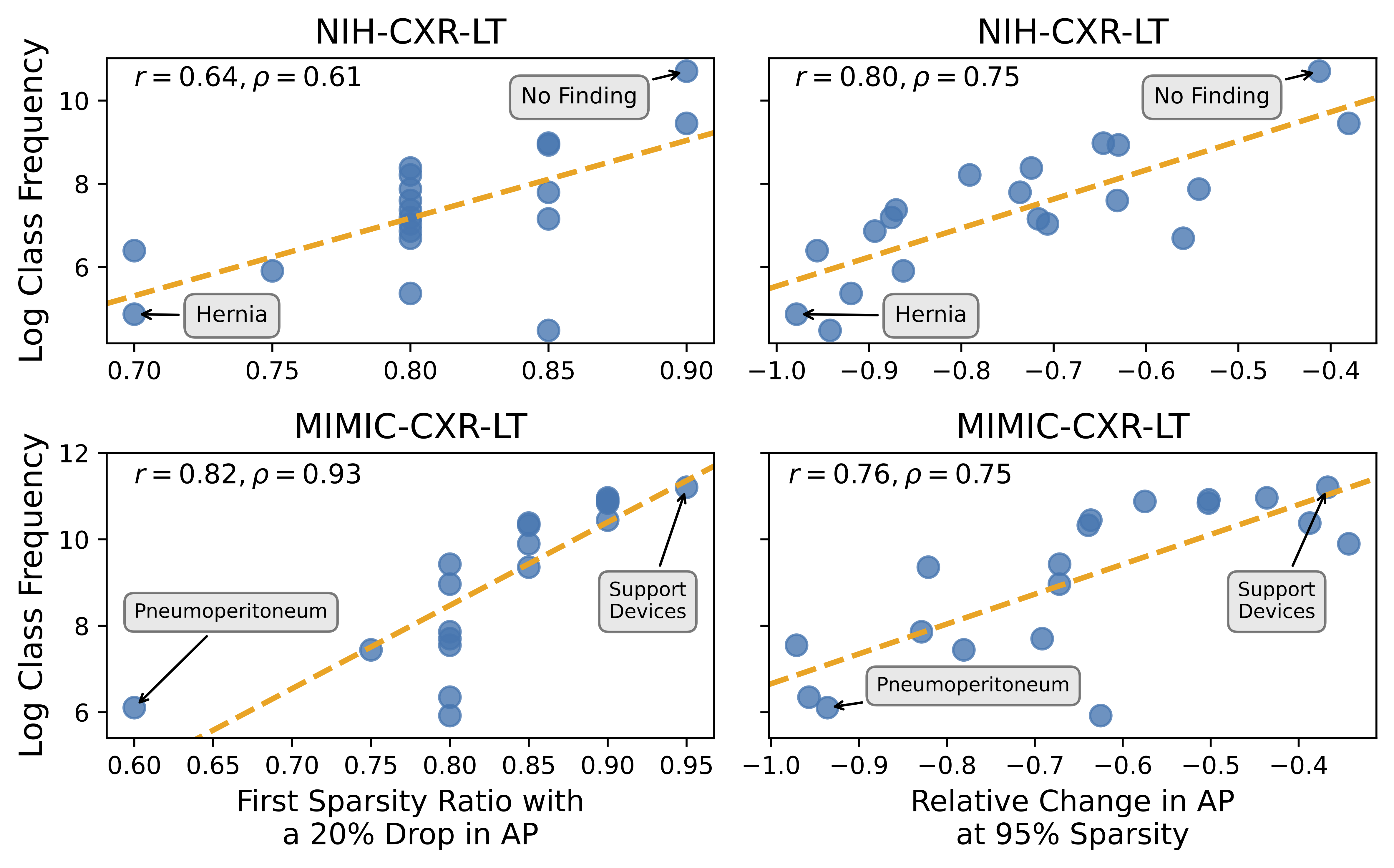
nih-cxr-lt_L1-prune_predsandmimic-cxr-lt_L1-prune_predsand place both directories in this repo's base directory. The script to perform L1 pruning and inference on all models trained in the previous step can be found insrc/prune.py. - Generate the content of Figures 1-4 by running
python src/main_analysis.py. - Generate the content of Figure 5 by running
python src/pie_analysis.py.
While the raw data from the radiologist reader study described in Section 2.3 is not publicly available, the code to generate Figure 6, which summarizes radiologist perception of PIEs, can be found in src/pie_survey_analysis.py.
Please either open a Github Issue or reach out to Greg Holste at gholste@utexas.edu with any questions.