In progress. Submitted to NeuroImage: Clinical.
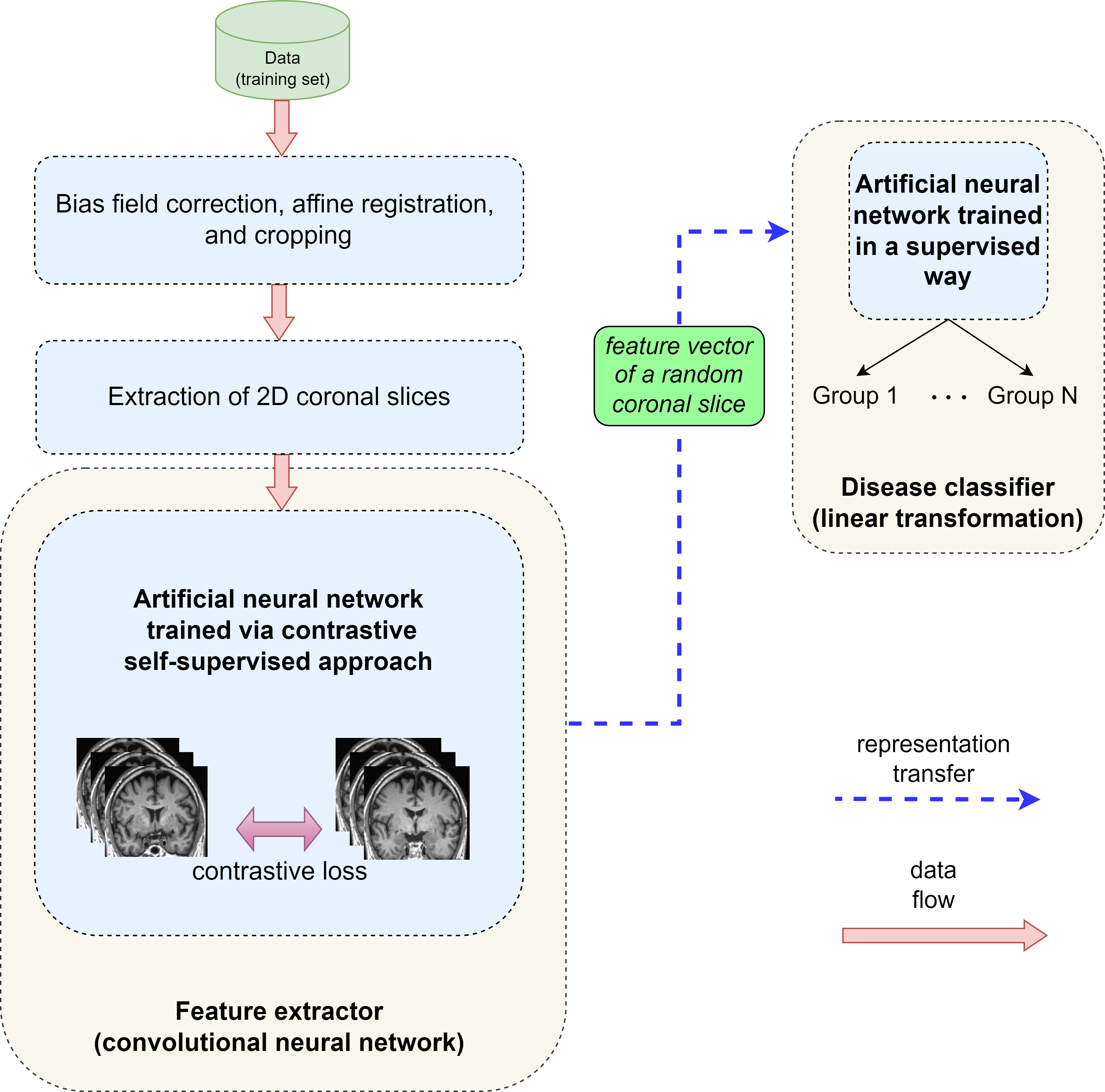
Explainable Differential Diagnosis of Dementia using Self-supervised Learning. The proposed method is based on Nearest-Neighbour Contrastive Learning of Visual Representations (NNCLR) [1] using ConvNeXt Tiny [2] with 7x7 filter kernels.
The following image visualizes the whole architecture:
configurationfolder contains the YAML file for the configurationdata_processingfolder contains the methods that are used to prepare data for a modelmodelsfolder defines a model structurerun_train_nnclr.pytrains the NNCLR modelrun_train-classifier.pytrains the classification modelrun_classifier_indep_eval.pyevaluates the whole model on independent datarun_classifier_evaluation.pyevaluates the whole model on the validation/test setrun_visualcreates figures- All output information will be saved into the working folder (see
configuration.yaml)
Datasets are accessible through http://adni.loni.usc.edu/, if not otherwise stated:
- (in use) ADNI. Alzheimer's Disease Neuroimaging Initiatve
- (in use) NIFD. Frontotemporal Lobar Degeneration
- (in use) AIBL. Australian Imaging, Biomarkers and Lifestyle
- (in use) OASIS. Access: https://www.oasis-brains.org/#data
- (planned) ABVIB. Aging Brain: Vasculature, Ischemia, and Behaviour
- (in use) PPMI. Parkinson's Progression Markers Initiative
- (planned) GCP. Brain Genomics Superstruct Project
- (planned) The A4 Study. Anti-Amyloid Treatment in Asymptomatic Alzheimer's
- (planned) MIRIAD, Access: https://www.ucl.ac.uk/drc/research/research-methods/minimal-interval-resonance-imaging-alzheimers-disease-miriad
- (planned) HABS, Access: will follow
See also /data_dzne_archiv2/Studien/ClinicNET/data/
sudo apt-get install dcm2niix- install ANTs: https://github.com/ANTsX/ANTs/wiki/Compiling-ANTs-on-Linux-and-Mac-OS
pip3 install captum lightly openpyxl plotly mlxtend zennit nibabel torch_tb_profilergit clone https://github.com/VadymV/clinicaandgit clone https://github.com/VadymV/clinicadl.gitpip install ./clinica ./clinicadlconda install tsnecuda -c conda-forge- download each dataset and place extracted image data into its own directory:
/data/dataset_name/data/folder_with_extracted_data (e.g. /data/nifd/data/NIFD).
Also download clinical data and place it into the clinical_data folder (e.g. /data/nifd/clinical_data).
Of course, a symbolic link can be used instead of using the dataset directory directly (e.g.
ln -s source link) - follow the instructions on https://aramislab.paris.inria.fr/clinica/docs/public/latest/
how to use dataset converters: e.g.
clinica convert nifd-to-bids [OPTIONS] DATASET_DIRECTORY CLINICAL_DATA_DIRECTORY BIDS_DIRECTORY. - run
clinica run t1-linear [OPTIONS] BIDS_DIRECTORY CAPS_DIRECTORYSee also https://aramislab.paris.inria.fr/clinica/docs/public/latest/Pipelines/T1_Linear/ for more details - extract 2D slices:
clinicadl extract slice CAPS_DIRECTORY t1-linear --slice_mode single --slice_direction 1 --discarded_slices 50 - create a file that contains all required information about patients:
clinica iotools merge-tsv BIDS info_data.tsv
- When data are prepared the actual training of models and their evaluation can begin
- The file
info_data.tsvis used as metadata (e.g. for data loading, split into train and test sets, creation of labels) - Configurations are defined in the
configuration/configuration.yamlfile:- Block
datacontains settings on how to select slices and what labels to consider nnclr,classifier, andindependent_evaluationblocks are used to set parameters for training/evaluation and load specific dataset
- Block
- Do you want to use your own dataset? Extend the code in
clinicaandclinicadllibraries:- Conversion to BIDS: see
clinica/clinica/iotools/convertersto imitate the same structure - T1-pipeline: see
clinica/clinica/pipelines/t1_linearif in some places additional references are needed, however the pipeline should not depend on additional information - Extraction of slices: see
clinicadl/clinicadl/extractif in some places additional references are needed, however the pipeline should not depend on additional information
- Conversion to BIDS: see
- NNCLR training:
ADNI3,ADNI2,NIFD - Classifier training:
ADNI3,ADNI2,NIFD - Training trials: 3 (each trial new train and test sets are created that are the same for NNCLR and classifier models)
- Independent evaluation:
AIBL - Sample statistics of ADNI3 (N=2365, Patients=844):
| CN | AD | MCI | |
|---|---|---|---|
| Age | 73.99(7) | 76.96(8.31) | 74.57(7.97) |
| MMSE | 29.38(0.73) | 20.84(4.50) | 27.85(1.10) |
| Sex: F/M | 312/221 | 52/70 | 140/173 |
- Sample statistics of ADNI2 (N=1836, Patients=743):
| CN | AD | MCI | |
|---|---|---|---|
| Age | 75.75(7.02) | 76.22(7.63) | 74.57(7.86) |
| MMSE | 29.31(0.74) | 21.07(4.29) | 27.75(1.12) |
| Sex: F/M | 110/94 | 120/163 | 151/203 |
- Sample statistics of AIBL (N=991, Patients=583):
| CN | AD | MCI | |
|---|---|---|---|
| Age | 73.51(6.41) | 75.39(7.86) | 76.62(6.52) |
| MMSE | 29.20(0.77) | 19.45(5.57) | 27.17(1.25) |
| Sex: F/M | 239/182 | 51/37 | 41/62 |
- Sample statistics of NIFD (N=614, Patients=273):
| CN | BV | SV | PNFA | |
|---|---|---|---|---|
| Age | 64.29(7.05) | 62.09(5.82) | 62.72(6.80) | 68.94(7.72) |
| MMSE | 29.68(0.47) | 22.56(6.22) | 22.48(5.74) | 24.92(5.50) |
| Sex: F/M | 72/58 | 23/48 | 14/23 | 19/16 |
-
Sample statistics of OASIS: N=2012, Patients=1079:
-
Sample statistics of OASIS: N=1411, Patients=835:
-
NNCLR training:
- 1,000 epochs
- a batch size of 180
- nearest neighbour size: 8192
- ConvNeXt Tiny CNN model with 7x7 filter kernel
- data preparation for each sample:
- a random slice across coronal plane within the middle region of the brain is selected
- random transformations
tare applied sequentially withp(t)=0.5to get 2 views of the same sample:- resized crop
- erasing
- horizontal flip
-
Training of a classifier:
- ConvNeXt Tiny CNN model serves as a feature extractor and is not trained
- 100 epochs
- a batch size of 64
- features dimension from ConvNeXt Tiny CNN model: 768
- classifier block that is trained: normalisation layer, flat operation, linear layer
-
Feature maps of 22 convolutional layers (subject ADNI002S0729 diagnosed with AD, session M60):
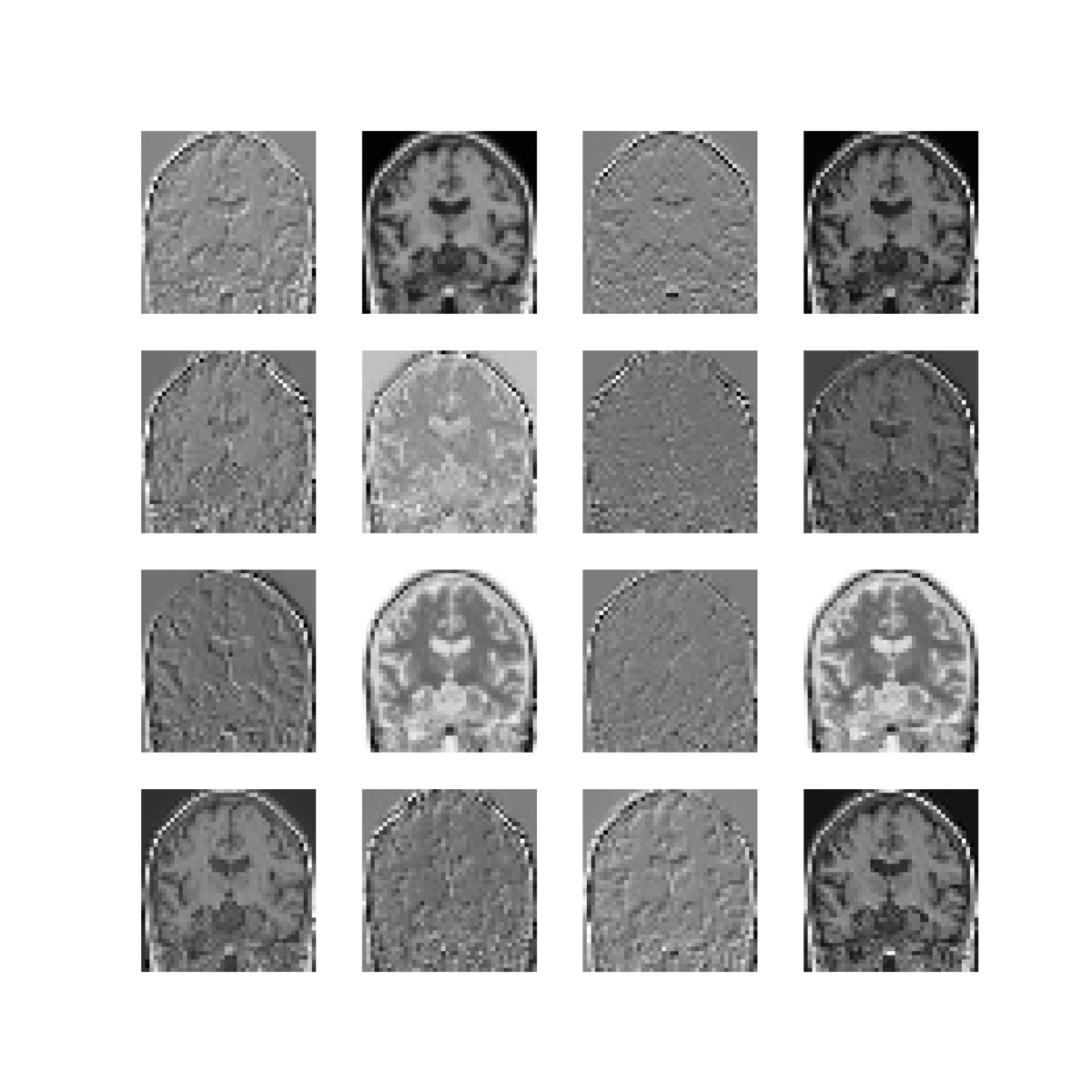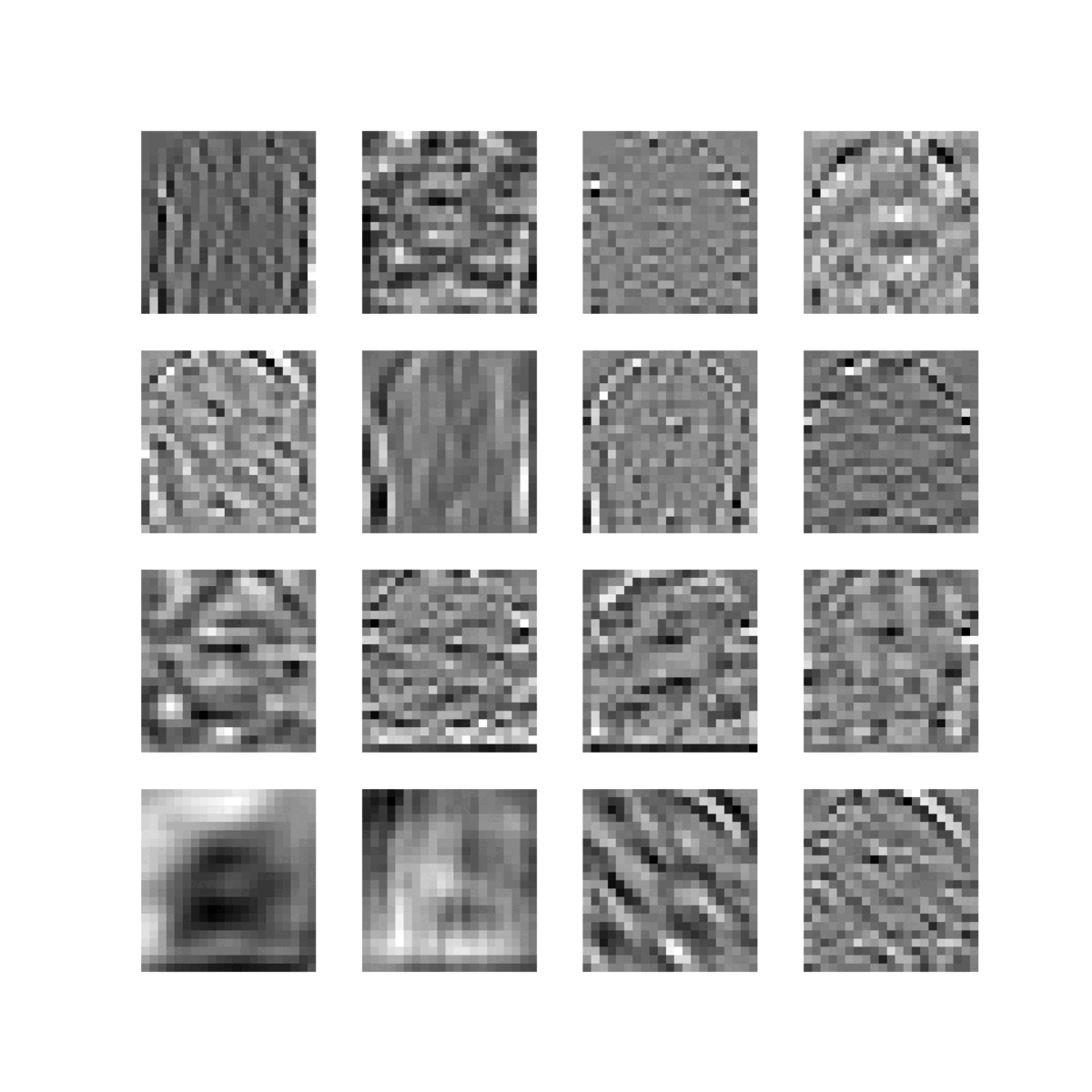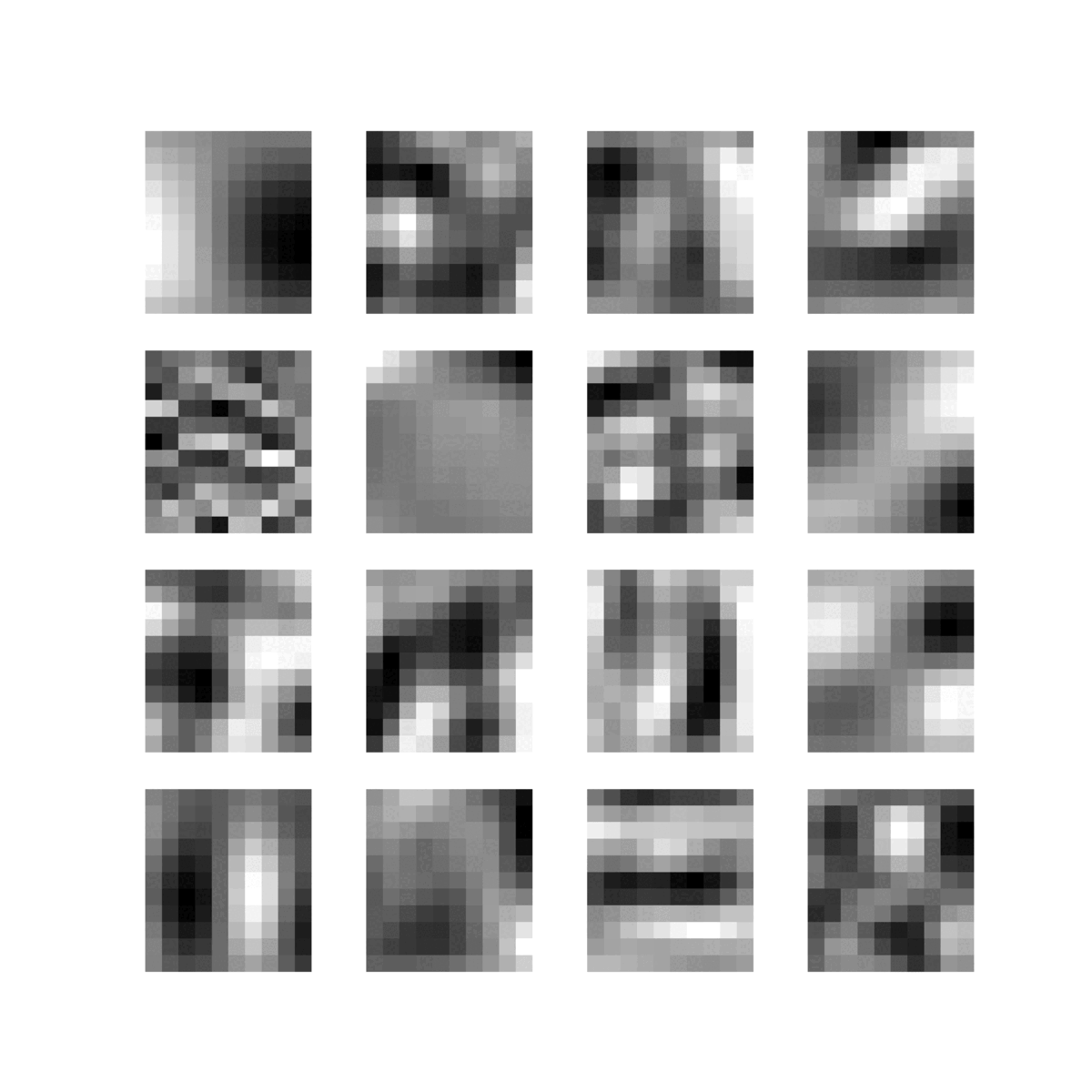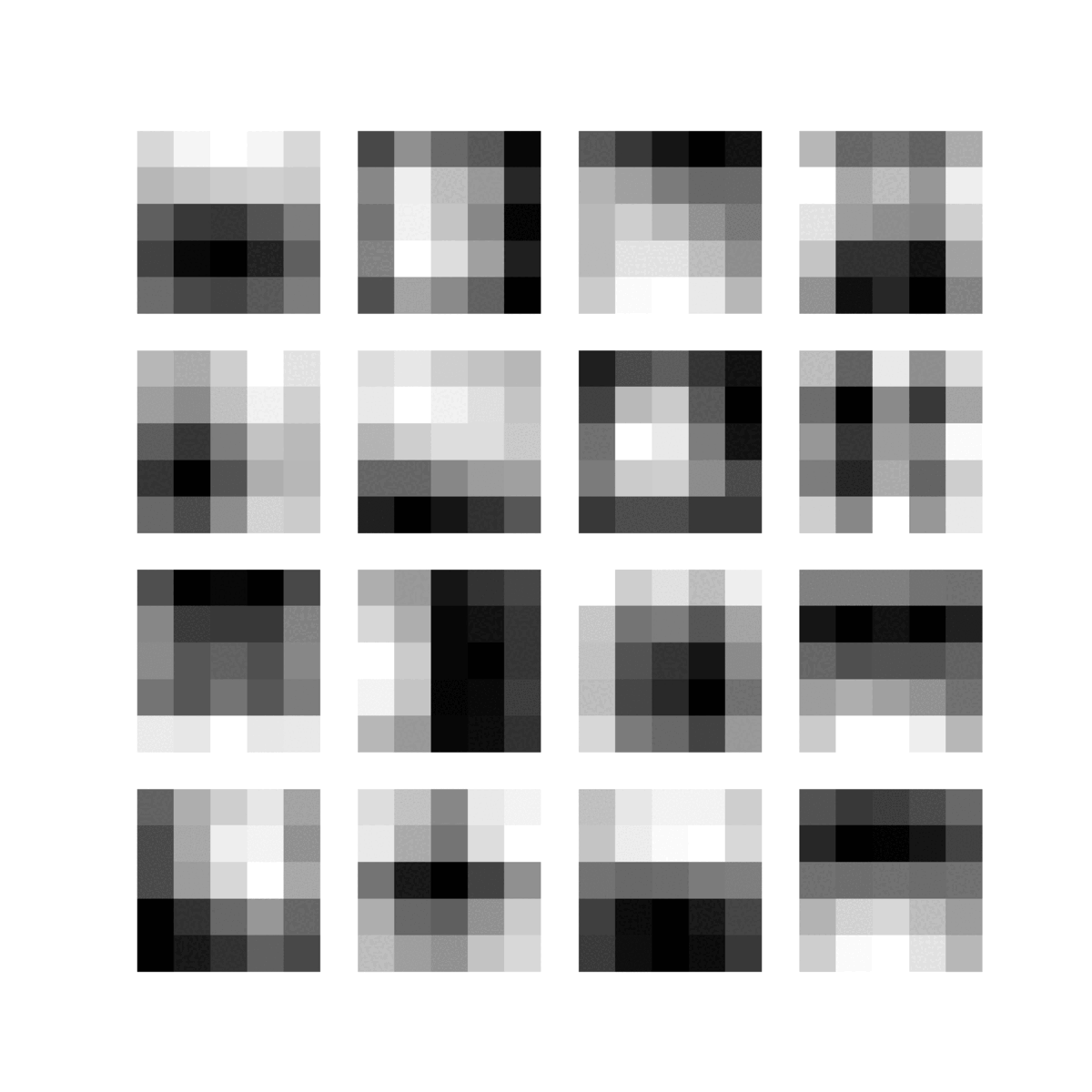
-
t-SNE visualisation of features learned by NNCLR (train set):
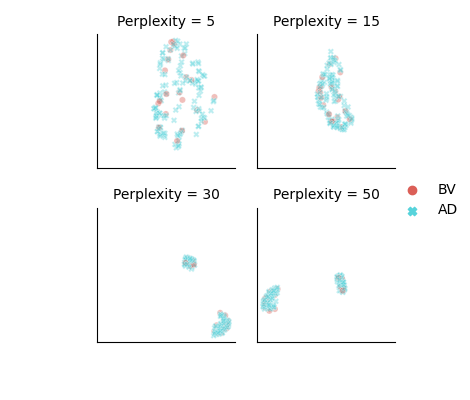
-
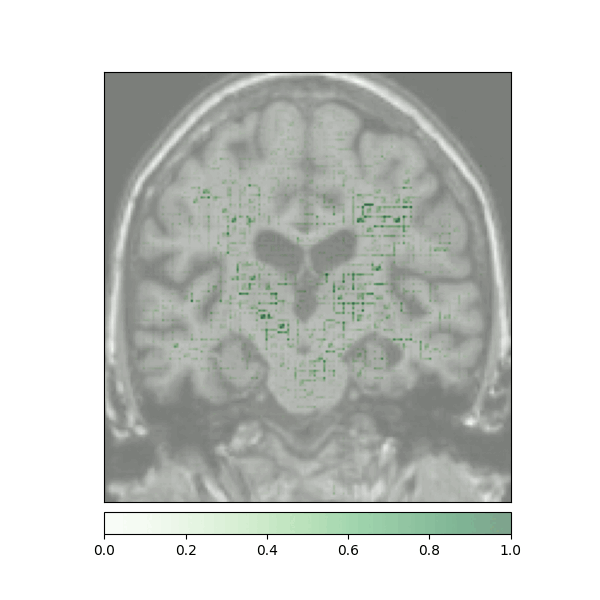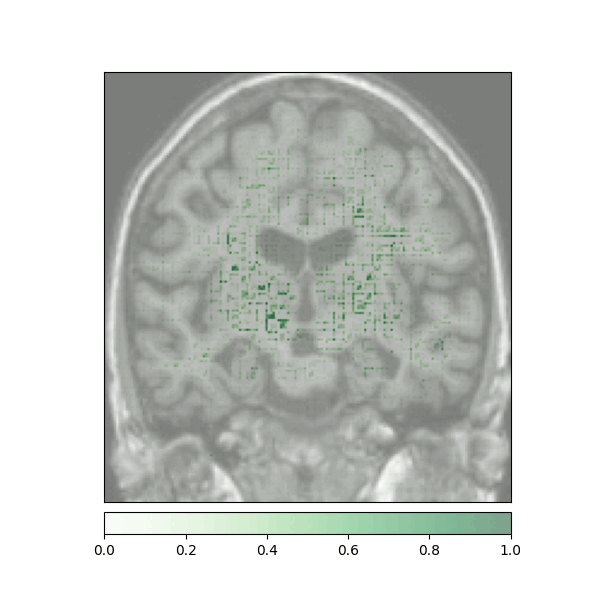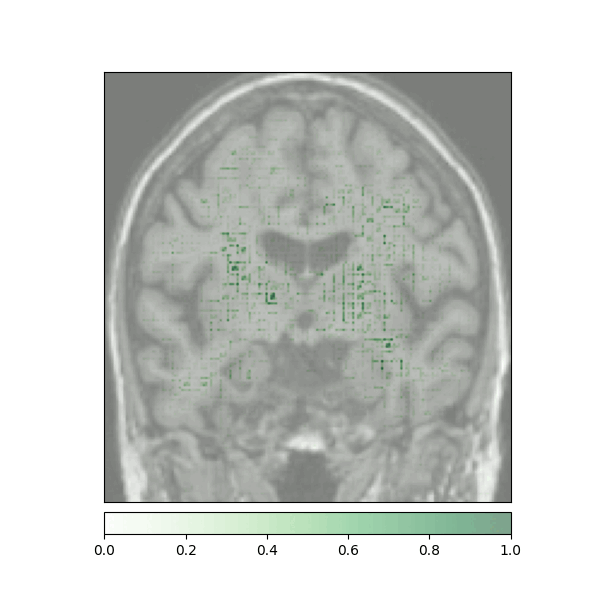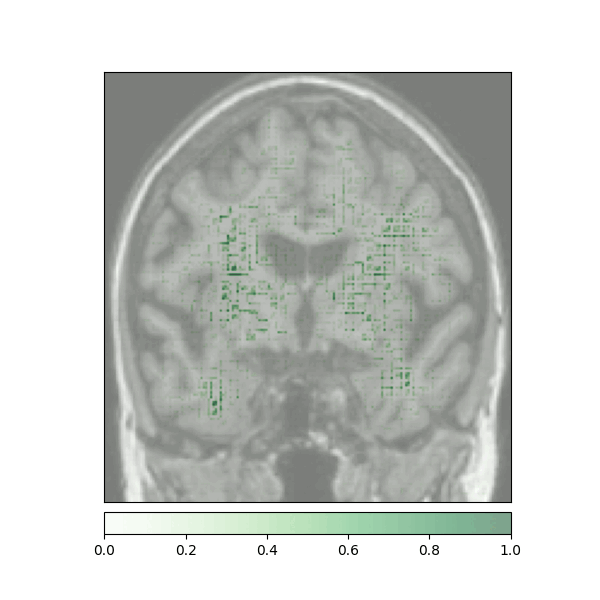
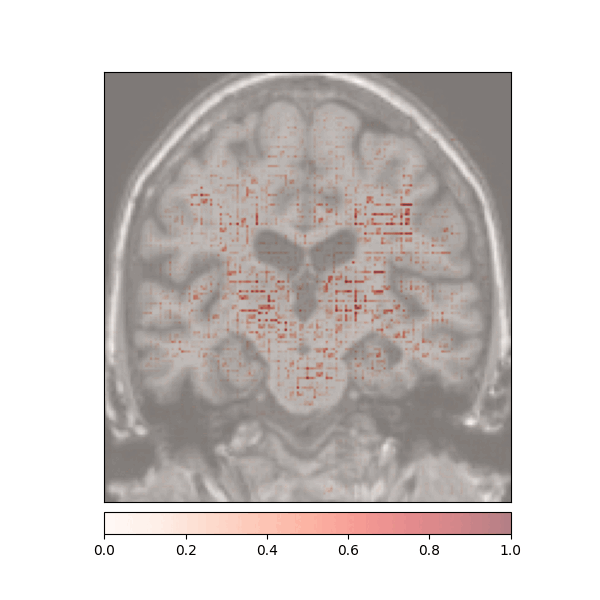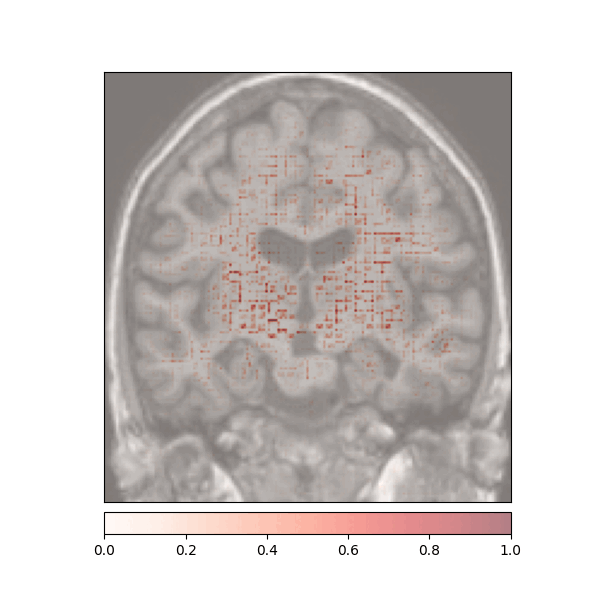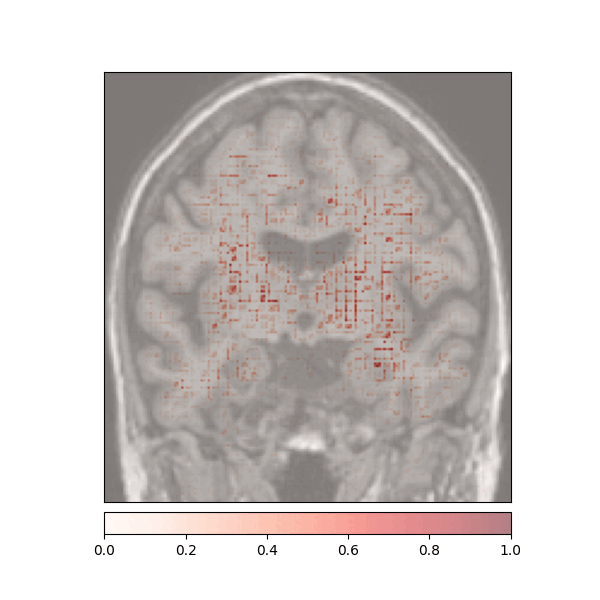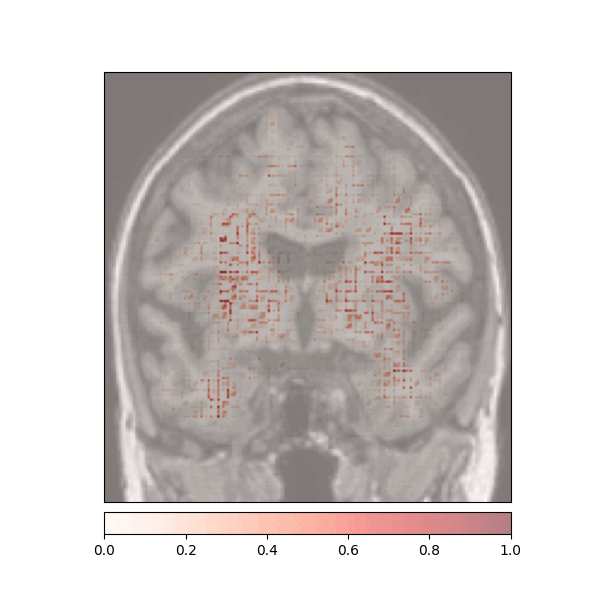
Attributions created using Integrated Gradients of a patient diagnosed with Alzheimer's Disease: green (positive atttributions), red (negative attributions)
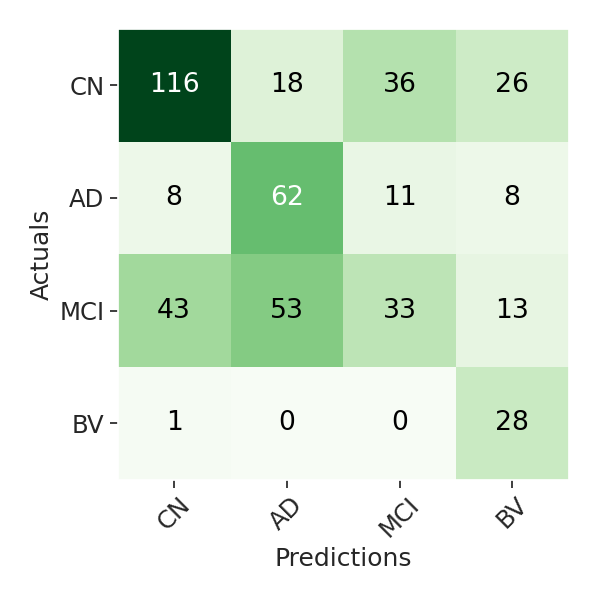
- Evaluation results:
[1] Dwibedi, D., Aytar, Y., Tompson, J., Sermanet, P., & Zisserman, A. (2021). With a Little Help From My Friends: Nearest-Neighbor Contrastive Learning of Visual Representations. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), 9588-9597.
[2] Liu, Z., Mao, H., Wu, C.Y., Feichtenhofer, C., Darrell, T., & Xie, S.. (2022). A ConvNet for the 2020s.
-
delete files:
find . -name '*pattern*' -exec rm {} \; -
unzip files:
unzip "file_name*.zip" -d output_dir/ -
running out of storage place. Clinica uses tmp directory for intermediate calculations. If you want to provide another location, do:
export TEMP=/new_location/orexport TMP=/new_location/orexport TMPDIR=/new_location/based on your settings -
clinicadl (version 1.0.3) requires clinica==0.4.1
-
if
InvalidIndexErroroccurs, insert (line 157):if not merged_df.empty: merged_df = merged_df.append(row_df) else: merged_df = row_df.copy() -
if an error
ValueError: cannot reindex from a duplicate axisoccurs insert after line 167:merged_df = merged_df.reset_index(drop=True) merged_df = merged_df.loc[:, ~merged_df.columns.duplicated()]
-
(NIFD) T1w MRI file for the subject sub-NIFD1S0017 with session ses-M00 does not exist and thus is removed
-
PyCharm does not have enough space: set the variables idea.system.path=/path idea.log.path=/data_dzne_archiv/Studien/ClinicNET/temp/log/
-
Warning: Assuming 7FE0,0010 refers to an icon not the main image. This warning was ignored. Upgrade of dcm2niix did not help. -
pandas.errors.ParserError: Expected 36 fields in line 80, saw 37. This error occurs while parsing the file 'aibl_flutemeta*.csv'. Fix: replace 'measured, AUSTIN AC AT Brain H19s' through 'measured AUSTIN AC AT Brain H19s'. -
File not founderror during the execution of the command ``xxxx-to-bids. To overcome the termination of the process, typecontinue` in a file `clinica -> utils -> inputs.py` just before where the errors are collected: line 309 (08.02.2022, clinica=0.5.3) -
dx1is instead ofdiagnosisas a column name during processing of the OASIS dataset. Insert the following into the fileclinica/clinica/iotools/utils/data_handling.pyat the appropriate places:
if 'dx1' in sessions_df.columns:
sessions_df.rename(
columns={
"dx1": "diagnosis",
}, inplace=True,
)