Implementation of ElasticSearch for the indexing, ranking, and retrieval of Drug Molecules to aid in In silico Drug Design
Usually molecules are stored/represented in a typical smiles format
Drug Discovery has become more and more expensive over the years. One of the most challenging parts is finding candidate drug molecules that will be optimized in later stages. While drug discovery is becoming more challenging, at the same time, datasets related to drug discovery are continually increasing in size and are now more accesible to the public. Examples include;
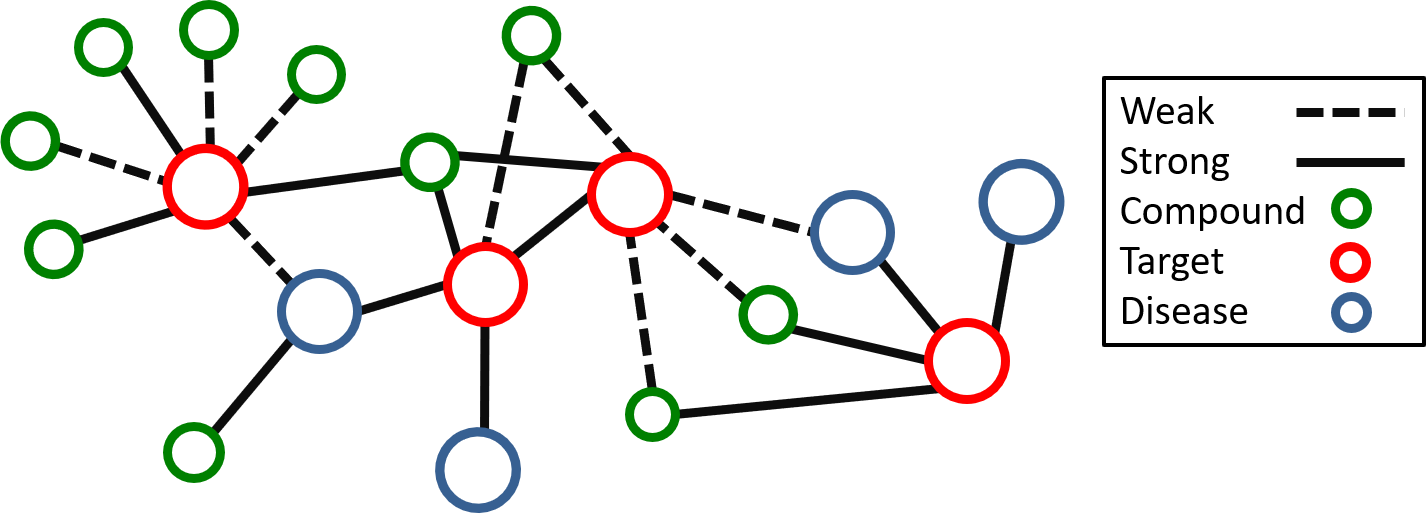
To reduce the cost of drug discovery, we can apply big data techniques to mine these chemical databases and find new uses for old drug molecules, underutilized protein targets, or novel pathways for diseases.
To help in mining these unstructured and messy datasets, I have created a python package to translate these datasets into drug interaction networks which act as graph databases. Using these graph databases, medicinal chemists can quickly search, filter, and prepare subsets of the drug interaction network related to their study. These smaller datasets can be used for machine learning models, visualizations, and property anlaysis. Drug interaction networks stored in graph databases will help streamline the drug discovery process by making the tedious task of dataset preparation much more simpler and user friendly.
Elasticsearch is an open-source, distrubuted, RESTful search and analytics engine that has been widely used in
python 3.6elasticsearchelasticsearch-dslrdkit