Updated: 2020-06-07
Synopsis
This is a data analysis report of the public available data about Covid19 in Portugal. The report demonstrates how to download and process data, ranks Portugal by cases and deaths and demonstrates the pandemic evolution in this country. Some final remarks are made about the limited availability of the data provided.
This report is also an example of reproducible research in data analysis making it possible to anyone to reproduce or adapt for any country.
Raw Data
World Data Source
The primarily data source used for this work was available by the European Centre for Disease Prevention and Control. By analyzing this data we can get a gist of the evolution of covid19 in the world. Some information were added trough time to this data, such as new formats. I choose the JSON format.
library(jsonlite)
library(dplyr)
url <- "https://opendata.ecdc.europa.eu/covid19/casedistribution/json"
dataRaw <- read_json(url, simplifyVector = TRUE)
data <- as_tibble(dataRaw$records)
str(data)
## Classes 'tbl_df', 'tbl' and 'data.frame': 21965 obs. of 11 variables:
## $ dateRep : chr "07/06/2020" "06/06/2020" "05/06/2020" "04/06/2020" ...
## $ day : chr "7" "6" "5" "4" ...
## $ month : chr "6" "6" "6" "6" ...
## $ year : chr "2020" "2020" "2020" "2020" ...
## $ cases : chr "582" "915" "787" "758" ...
## $ deaths : chr "18" "9" "6" "24" ...
## $ countriesAndTerritories: chr "Afghanistan" "Afghanistan" "Afghanistan" "Afghanistan" ...
## $ geoId : chr "AF" "AF" "AF" "AF" ...
## $ countryterritoryCode : chr "AFG" "AFG" "AFG" "AFG" ...
## $ popData2018 : chr "37172386" "37172386" "37172386" "37172386" ...
## $ continentExp : chr "Asia" "Asia" "Asia" "Asia" ...
World Data Pre-Processing - getting Tiddy
Since all available variables are in character format I transformed the numeric values and date values into their respective formats.
library(lubridate)
data$dateRep <- dmy(data$dateRep)
data$day <- as.numeric(data$day)
data$month <- as.numeric(data$month)
data$year <- as.numeric(data$year)
data$cases <- as.numeric(data$cases)
data$deaths <- as.numeric(data$deaths)
data$popData2018 <- as.numeric(data$popData2018)
str(data)
## Classes 'tbl_df', 'tbl' and 'data.frame': 21965 obs. of 11 variables:
## $ dateRep : Date, format: "2020-06-07" "2020-06-06" ...
## $ day : num 7 6 5 4 3 2 1 31 30 29 ...
## $ month : num 6 6 6 6 6 6 6 5 5 5 ...
## $ year : num 2020 2020 2020 2020 2020 2020 2020 2020 2020 2020 ...
## $ cases : num 582 915 787 758 759 545 680 866 623 580 ...
## $ deaths : num 18 9 6 24 5 8 8 3 11 8 ...
## $ countriesAndTerritories: chr "Afghanistan" "Afghanistan" "Afghanistan" "Afghanistan" ...
## $ geoId : chr "AF" "AF" "AF" "AF" ...
## $ countryterritoryCode : chr "AFG" "AFG" "AFG" "AFG" ...
## $ popData2018 : num 37172386 37172386 37172386 37172386 37172386 ...
## $ continentExp : chr "Asia" "Asia" "Asia" "Asia" ...
Now we can work with these numbers and make some exploratory analysis. Let’s check out a summary of our data:
summary(data)
## dateRep day month year
## Min. :2019-12-31 Min. : 1.00 Min. : 1.000 Min. :2019
## 1st Qu.:2020-03-18 1st Qu.: 7.00 1st Qu.: 3.000 1st Qu.:2020
## Median :2020-04-16 Median :16.00 Median : 4.000 Median :2020
## Mean :2020-04-08 Mean :15.73 Mean : 3.817 Mean :2020
## 3rd Qu.:2020-05-12 3rd Qu.:24.00 3rd Qu.: 5.000 3rd Qu.:2020
## Max. :2020-06-07 Max. :31.00 Max. :12.000 Max. :2020
##
## cases deaths countriesAndTerritories
## Min. :-2461.0 Min. :-1918.00 Length:21965
## 1st Qu.: 0.0 1st Qu.: 0.00 Class :character
## Median : 3.0 Median : 0.00 Mode :character
## Mean : 311.2 Mean : 18.15
## 3rd Qu.: 52.0 3rd Qu.: 1.00
## Max. :48529.0 Max. : 4928.00
##
## geoId countryterritoryCode popData2018 continentExp
## Length:21965 Length:21965 Min. :1.000e+03 Length:21965
## Class :character Class :character 1st Qu.:2.083e+06 Class :character
## Mode :character Mode :character Median :9.485e+06 Mode :character
## Mean :4.888e+07
## 3rd Qu.:3.199e+07
## Max. :1.393e+09
## NA's :326
Ok, something awkward is going on. The new cases and deaths variable have negative values, because, as one may notice, the minimum value is -2461 and -1918 for each. There shouldn’t be negative values, the only explanation available is mistakes in counts that are then corrected this way. This is possible the wrongest way to correct mistakes because one doesn’t know for sure the real values of the mistaken day nor the negative cases/deaths day.
One thing to always have in mind is the source of the data: our source must be reliable and trustworthy.
Now that I have a somewhat tidy data, I can think a bit about the variables available:
names(data)
## [1] "dateRep" "day"
## [3] "month" "year"
## [5] "cases" "deaths"
## [7] "countriesAndTerritories" "geoId"
## [9] "countryterritoryCode" "popData2018"
## [11] "continentExp"
The most interesting data I will address will be the number of detected cases and deaths, by date reported.
Portugal in the World
How is Portugal rated in death and cases counts? There is some social media discussion about how to address this rates. Let us compare absolute numbers with percentages related to country population:
CasesRank <- data %>%
group_by(countriesAndTerritories) %>%
summarise("TotalCases" = sum(cases)) %>%
arrange(desc(TotalCases))
PTrankCases <- grep("Portugal", CasesRank$countriesAndTerritories)
numberofCases <- CasesRank %>% filter(countriesAndTerritories == "Portugal")
numberofCases <- as.integer(numberofCases[2])
knitr::kable(head(CasesRank, 10))
| countriesAndTerritories | TotalCases |
|---|---|
| United_States_of_America | 1920061 |
| Brazil | 645771 |
| Russia | 458689 |
| United_Kingdom | 284868 |
| India | 246628 |
| Spain | 241310 |
| Italy | 234801 |
| Peru | 191758 |
| Germany | 183979 |
| Iran | 169425 |
The above table reflects the ranked 10 worst countries in COVID-19 diagnosed cases. Portugal is ranked 30, with 34351 total cases.
DeathsRank <- data %>%
group_by(countriesAndTerritories) %>%
summarise("TotalDeaths" = sum(deaths)) %>%
arrange(desc(TotalDeaths))
PTrankDeaths <- grep("Portugal", DeathsRank$countriesAndTerritories)
numberofDeaths <- DeathsRank %>% filter(countriesAndTerritories == "Portugal")
numberofDeaths <- as.integer(numberofDeaths[2])
knitr::kable(head(DeathsRank, 10))
| countriesAndTerritories | TotalDeaths |
|---|---|
| United_States_of_America | 109802 |
| United_Kingdom | 40465 |
| Brazil | 35026 |
| Italy | 33846 |
| France | 29142 |
| Spain | 27135 |
| Mexico | 13511 |
| Belgium | 9580 |
| Germany | 8668 |
| Iran | 8209 |
The above table reflects the ranked 10 worst countries in COVID-19 deaths. Portugal is ranked 25, with 1474 total deaths.
Let us now check the same rates in percentage.
CasesPerRank <- data %>%
group_by(countriesAndTerritories) %>%
summarise("TotalCases" = sum(cases), "Pop" = unique(popData2018)) %>%
mutate("PercentageCases" = TotalCases/Pop * 100) %>%
arrange(desc(PercentageCases))
DeathsPerRank <- data %>%
group_by(countriesAndTerritories) %>%
summarise("TotalDeaths" = sum(deaths), "Pop" = unique(popData2018)) %>%
mutate("PercentageDeaths" = TotalDeaths/Pop * 100) %>%
arrange(desc(PercentageDeaths))
PTPerRankCases <- grep("Portugal", CasesPerRank$countriesAndTerritories)
PTPerRankDeaths <- grep("Portugal", CasesPerRank$countriesAndTerritories)
knitr::kable(head(CasesPerRank, 10))
| countriesAndTerritories | TotalCases | Pop | PercentageCases |
|---|---|---|---|
| Cases_on_an_international_conveyance_Japan | 696 | 3000 | 23.2000000 |
| Qatar | 67195 | 2781677 | 2.4156291 |
| San_Marino | 695 | 33785 | 2.0571259 |
| Holy_See | 12 | 1000 | 1.2000000 |
| Andorra | 852 | 77006 | 1.1064073 |
| Bahrain | 14383 | 1569439 | 0.9164421 |
| Kuwait | 31131 | 4137309 | 0.7524456 |
| Chile | 127745 | 18729160 | 0.6820648 |
| Singapore | 37527 | 5638676 | 0.6655286 |
| Luxembourg | 4035 | 607728 | 0.6639483 |
knitr::kable(head(DeathsPerRank, 10))
| countriesAndTerritories | TotalDeaths | Pop | PercentageDeaths |
|---|---|---|---|
| Cases_on_an_international_conveyance_Japan | 7 | 3000 | 0.2333333 |
| San_Marino | 42 | 33785 | 0.1243155 |
| Belgium | 9580 | 11422068 | 0.0838727 |
| Andorra | 51 | 77006 | 0.0662286 |
| United_Kingdom | 40465 | 66488991 | 0.0608597 |
| Spain | 27135 | 46723749 | 0.0580754 |
| Italy | 33846 | 60431283 | 0.0560074 |
| Sweden | 4656 | 10183175 | 0.0457225 |
| France | 29142 | 66987244 | 0.0435038 |
| Sint_Maarten | 15 | 41486 | 0.0361568 |
There are evident differences from percentage to absolute numbers in the extremes (but in fact Portugal doesn’t change much at the time of this report writing). In percentage Portugal is in 31 place for cases and 31 for deaths. Is this a fair comparison? There may be missing variables to understand our data: some index of number of urban centers per country for example, and also the predominance of respiratory diseases, atmospheric pollution and elderly people percentage. Also, the number of tests each country do highly influence the reported cases. All this summed together can cause a great impact at the political level for managing the emergency states of each countries and interpreting the results. With too many unknown factors the effectiveness of some policies may result only by chance.
Since there are just too many unknown factors in the percentage rate maybe the fairest way to compare is the absolute numbers (aware of not being ideal as well).
Evolution of the Disease in Portugal and Simulations
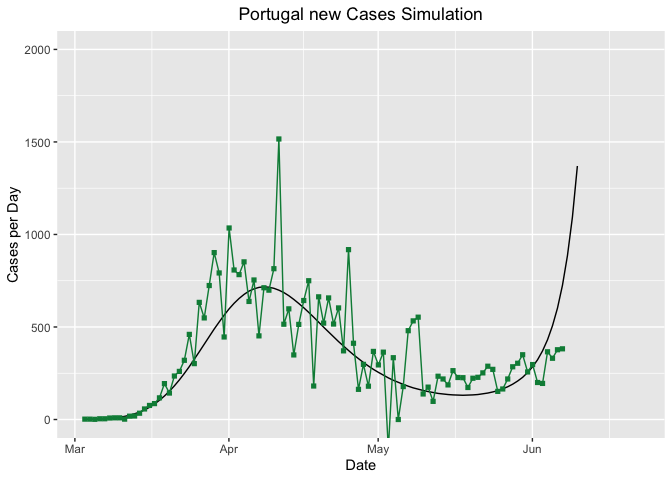
In the next lines I try to simulate new cases and deaths from COVID-19 in Portugal with a 15 day advance from the last data reported and to identify a peak, where we could consider a turning point for the pandemic in Portugal, meaning that the contingency politics are taking effect.
PTdata <- filter(data, geoId == "PT")
PTdataArranged <- arrange(PTdata, dateRep)
library(ggpmisc)
x <- 1:length(PTdataArranged$dateRep)
y <- log10(PTdataArranged$cases)
y <- gsub("[-InfNaN]", 0, y)
xsq <- x^2
xcub <- x^3
fit <- lm(y~x+xsq+xcub)
xv <- seq(min(x), 100, 1)
yv <- predict(fit, list(x = xv, xsq = xv^2, xcub = xv^3))
#Prediction <- tibble(Day = xv, logCases = yv)
PredictionCases <- tibble(Day = as.Date("2020-03-02")+xv,
SimCases = as.integer(10^yv),
RealCases = c(PTdataArranged$cases, rep(NA,
100-length(PTdataArranged$cases))))
PredictionCases$Day <- as.POSIXct(PredictionCases$Day)
CasesMaxDay <- as.Date(PredictionCases$Day[
grep(max(PredictionCases$SimCases[1:length(PTdataArranged$cases)]),
PredictionCases$SimCases)])
curvePredict <- ggplot(PredictionCases, aes(Day, SimCases))
PTgSimCasesNEW <- curvePredict + geom_line() +
geom_point(aes(Day, RealCases), col = "springgreen4", pch = 15) +
geom_line(aes(Day, RealCases), col = "springgreen4") +
labs(y = "Cases per Day", x = "Date",
title = "Portugal new Cases Simulation") +
stat_peaks(col = "tomato3", ignore_threshold = .9) +
theme(plot.title = element_text(hjust = 0.5)) +
coord_cartesian(xlim = c(PredictionCases$Day[1], as.POSIXct(Sys.Date() + 15)),
ylim = c(0, 2000))
PTgSimCasesNEW
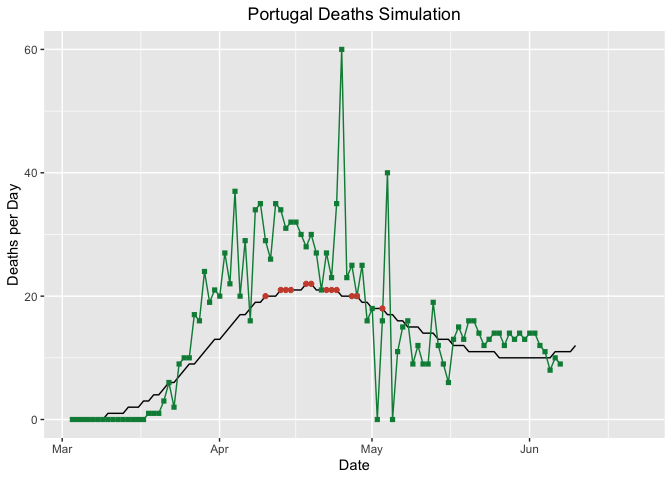
z <- log10(PTdataArranged$deaths)
z <- sub("-Inf", "0", z)
fitD <- lm(z~x+xsq+xcub)
xv <- seq(min(x), 100, 1)
zv <- predict(fitD, list(x = xv, xsq = xv^2, xcub = xv^3))
PredictionDeaths <- tibble(Day = as.Date("2020-03-02")+xv,
SimDeaths = as.integer(10^zv),
RealDeaths = c(PTdataArranged$deaths, rep(NA,
100-length(PTdataArranged$deaths))))
PredictionDeaths$Day <- as.POSIXct(PredictionDeaths$Day)
DeathsMaxDay <- as.Date(PredictionDeaths$Day[
grep(max(PredictionDeaths$SimDeaths[1:length(PTdataArranged$deaths)]),
PredictionDeaths$SimDeaths)])
curvePredictDeaths <- ggplot(PredictionDeaths, aes(Day, SimDeaths))
PTgSimDeaths <- curvePredictDeaths + geom_line() +
geom_point(aes(Day, RealDeaths), col = "springgreen4", pch = 15) +
geom_line(aes(Day, RealDeaths), col = "springgreen4") +
labs(y = "Deaths per Day", x = "Date",
title = "Portugal Deaths Simulation") +
stat_peaks(col = "tomato3", ignore_threshold = .7) +
theme(plot.title = element_text(hjust = 0.5)) +
coord_cartesian(xlim = c(PredictionDeaths$Day[1], as.POSIXct(Sys.Date() + 15)))
PTgSimDeaths
As we can observe from the graphs, the worst seems to have passed. The peak for new cases detected was in 2020-06-07 and the peak for deaths in Portugal was in 2020-04-18, 2020-04-19.
Conclusions
- Cases: Portugal is ranked 30 in 207 countries, with 34351 total cases. Portugal had the peak of cases in 2020-06-07.
- Deaths: Portugal is ranked 25 in 207 countries, with 1474 total deaths. Portugal had the peak of deaths in 2020-04-18, 2020-04-19.
The fact that the “peaks” have passed for the cases and deaths in
Portugal doesn’t mean that the problem is over. For example, the peaks
are always moving around because every day the simulations are iterated
with the new information available, and new policies and the end of the
confinement may produce more peaks for new cases and deaths.
From comparing our country with the others, we are quite better than
most countries. This is due to several factors, not only policies but
also with our peripheric geo-location. Also, it must be observed the
predominance of the disease in the north hemisphere of the planet - so
our rank my improve greatly for the next months (our rank in the end of
April is about 18th to 19th place), if there is some influence with the
flu-season.
Final Considerations
There is a lack of trustworthy data sources that would complement this data and give us detailed information about the how’s and why’s of SARS-CoV-2 behavior. Interesting variables that I would consider worth studying would be: Active Cases, Recovered Cases, ICU Cases, Non-ICU Hospitalized Cases, and also some information about patients, like: Known Diseases, a logical or more informative IfSmoker variable and Air Pollution Exposure.
Some of this information is retained by the “Sistema nacional de vigilância epidemiológica” SINAVE
I’ve applied for access in 2020-04-27 from an institutional e-mail but not a reply until today.