Community Detection
A library for Community Detection in graphs.
Introduction
Community Detection (or Community Search) is the process of finding sets of densely connected nodes in a graph which are structurally close to each other. We use the following algorithms in this library:
- Spectral Clustering
- Louvain Method
- Girvan-Newman algorithm
Application
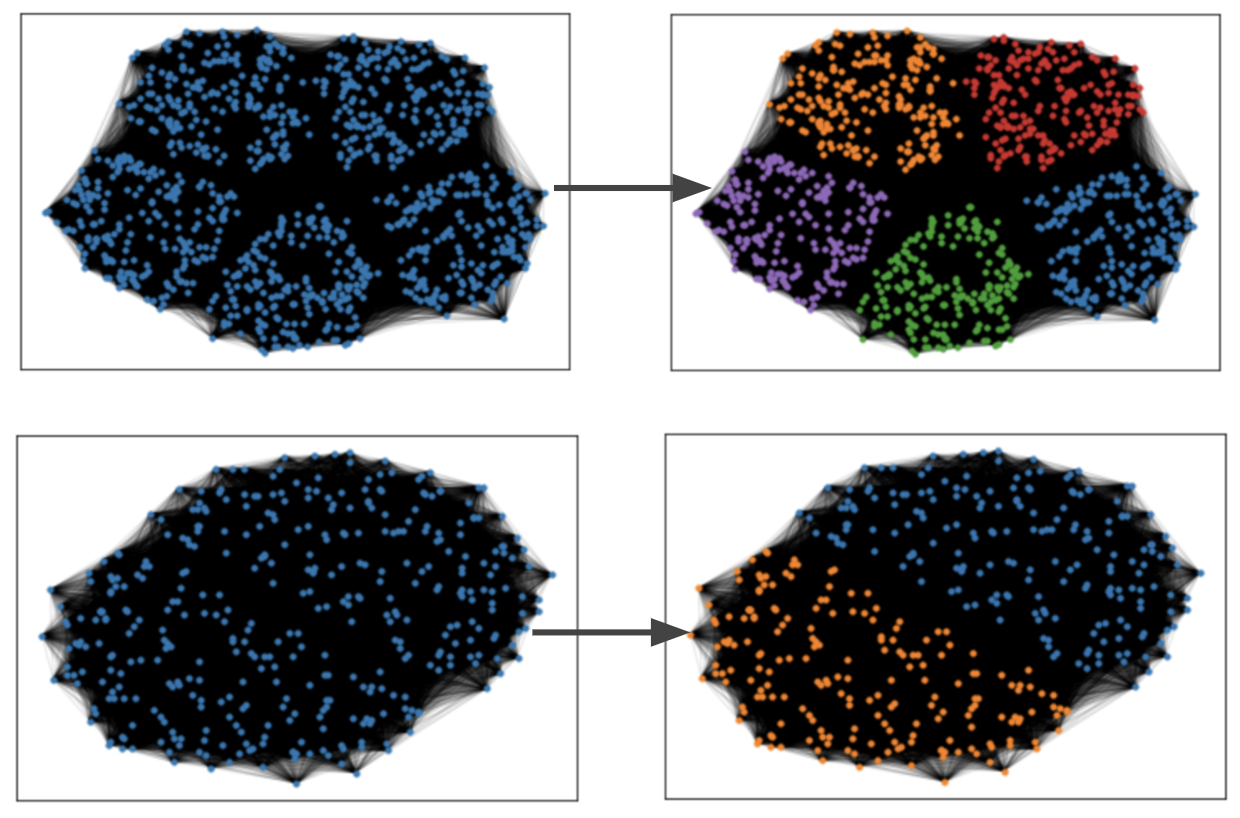
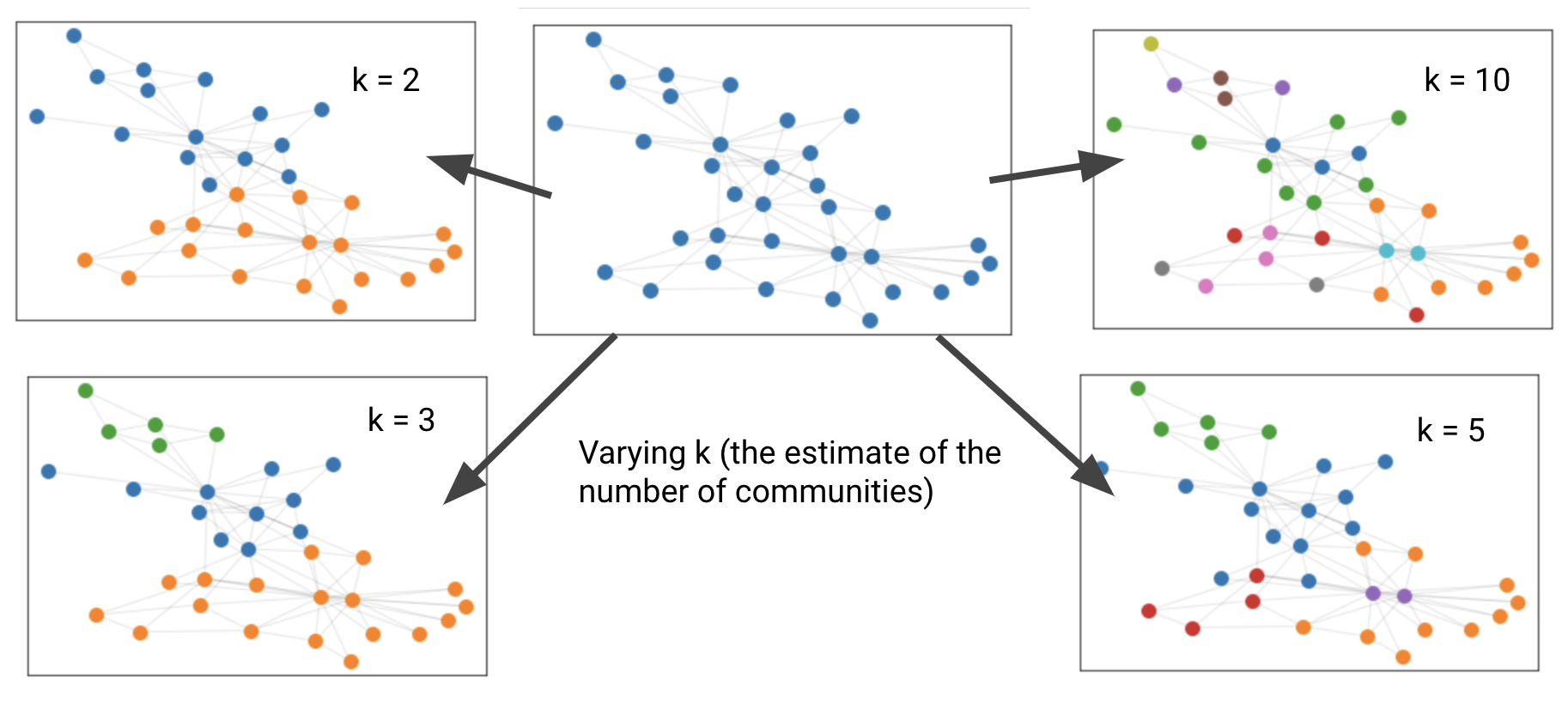
We tested our library on the Planted L-Partition Model (specifications in code) and Zachary’s Karate Club Network to visualize the results.
Planted L-Partition Model
Zachary’s Karate Club Network
Algorithms and Usage
Spectral Clustering
Test on Planted L Partition Model
import networkx as nx
from cdet.spectral_clustering import visualize_graph, spectral_clustering
# Color scheme
COLORS = \
["tab:blue", "tab:orange", "tab:green",
"tab:red", "tab:purple", "tab:brown",
"tab:pink", "tab:gray", "tab:olive",
"tab:cyan"]
# Create Planted L-Partition graph
K = 5
NODES_PER_BUCKET = 200
P_IN = 0.8
P_OUT = 0.1
G_pl = nx.generators.community.planted_partition_graph(K,
NODES_PER_BUCKET,
P_IN,
P_OUT)
pos_pl = nx.spring_layout(G_pl)
# Visualize base graph
visualize_graph(G_pl, pos_pl, edge_alpha=0.1, node_size=10, labels=False)
# Use Spectral Clustering and visualize
labels_dict = spectral_clustering(G_pl,
K,
pos_pl,
COLORS,
laplacian_type="symmetric",
edge_alpha=0.1,
node_size=10,
labels=False)Test on Zachary’s Karate Club Network
# Generate Zachary's Karate Club graph
G_kk = nx.karate_club_graph()
pos_kk = nx.spring_layout(G_kk)
# Visualize base graph
visualize_graph(G_kk, pos_kk)
# Use Spectral clustering and visualize
labels_dict = spectral_clustering(G_kk,
3,
pos_kk,
COLORS,
laplacian_type="symmetric")Louvain Method
Test on Planted L-Partition Model
import networkx as nx
from cdet.louvains import visualize_graph, louvains_method, plantedl
# Color scheme
COLORS = \
["tab:blue", "tab:orange", "tab:green",
"tab:red", "tab:purple", "tab:brown",
"tab:pink", "tab:gray", "tab:olive",
"tab:cyan"]
result = plantedl()
G = result[0]
pos = nx.spring_layout(G)
final_partition = result[1][0]
final_modularity = result[1][1]
labels_dict = {i: final_partition[i] for i in range(len(final_partition))}
# Visualize clustered graph
visualize_graph(G, pos, labels_dict=labels_dict, colors=COLORS, node_size=10)Girvan-Newman
You can use the original Networkx visualization options to visualize the outcomes from the algorithms as well.
Test on Planted L-Partition Model
# Original Planted L-Partition Graph
G = nx.planted_partition_graph(5, 30, 0.8, 0.1)
pos = nx.spring_layout(G, k=0.1, iterations=30, scale=1.3)
# Visualize using networkx API
nx.draw_networkx_nodes(G, pos=pos)
nx.draw_networkx_labels(G, pos=pos)
nx.draw_networkx_edges(G, pos=pos, width=2, alpha=1, edge_color='k')
plt.show()
# Planted L-Partition Graph graph with 2 communities
visualize_community_structure(num_communities=2, G)