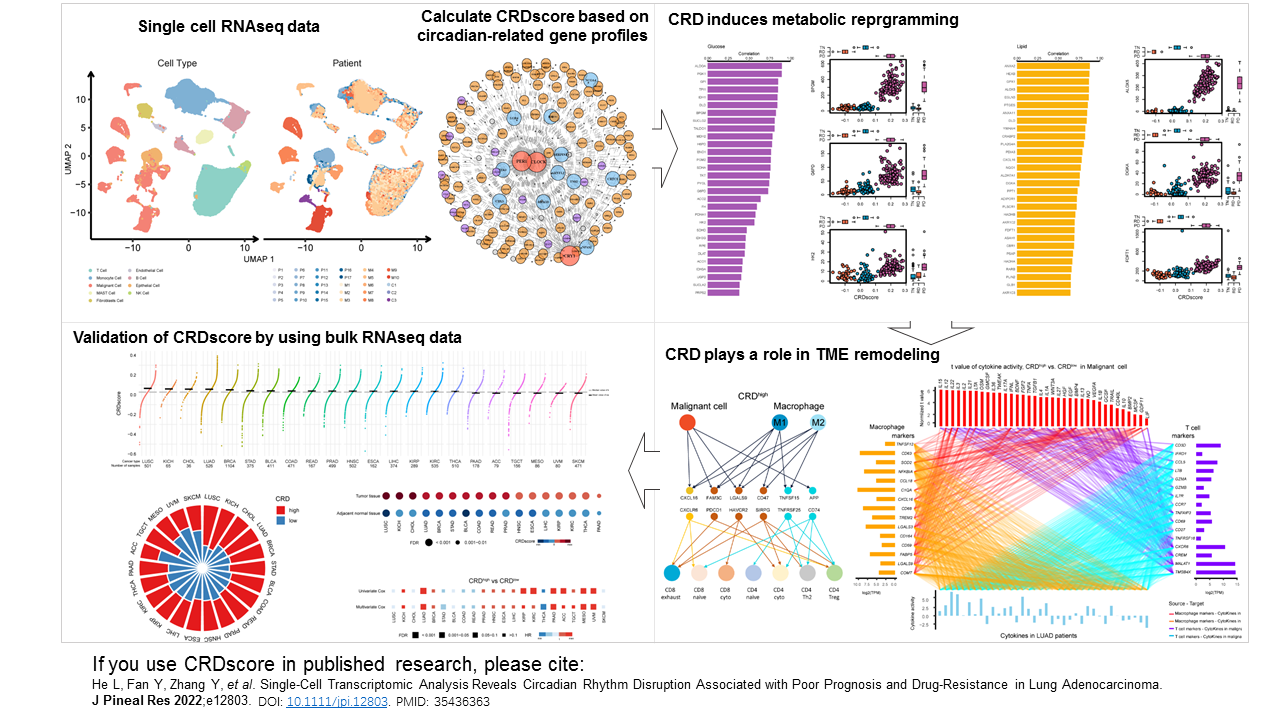
Circadian rhythm disruption (CRD) represents a major contributor to tumor progression, it plays
a pivotal role in metabolic reprogramming and T cell exhaustion.
The goal of CRDscore is to provide a novel computational framework for characterizing CRD status
using single-cell or bulk transcriptome data.
if (!require("devtools"))
install.packages("devtools")
devtools::install_github("yixianfan/CRDscore")
library(CRDscore)
data("exprset")
data("circadians_selected")
dim(exprset); head(exprset[,1:5])
head(circadians_selected); length(circadians_selected)
CRDscore <- cal_CRDscore(expr = exprset, n.bins = 50, circadians = circadians_selected, study.type = "scRNAseq")
head(CRDscore)
# graphic
library(tidyverse)
names(CRDscore) %>%
str_match(".*?_(.*?)_.*") %>%
as.data.frame() %>%
rename(sample_id = V1, gr = V2) %>%
mutate(CRDscore = CRDscore) %>%
ggplot(aes(x = gr, y = CRDscore, color = gr)) +
geom_boxplot()
If you use CRDscore in published research, please cite: He L, Fan Y, Zhang Y, et al. Single-Cell Transcriptomic Analysis Reveals Circadian Rhythm Disruption Associated with Poor Prognosis and Drug-Resistance in Lung Adenocarcinoma. J Pineal Res 2022;e12803. DOI: 10.1111/jpi.12803. PMID: 35436363