The Element Embeddings package provides high-level tools for analysing elemental and ionic species embeddings data. This primarily involves visualising the correlation between embedding schemes using different statistical measures.
- Documentation: https://wmd-group.github.io/ElementEmbeddings/
- Examples: https://github.com/WMD-group/ElementEmbeddings/tree/main/examples
Machine learning approaches for materials informatics have become increasingly widespread. Some of these involve the use of deep learning techniques where the representation of the elements is learned rather than specified by the user of the model. While an important goal of machine learning training is to minimise the chosen error function to make more accurate predictions, it is also important for us material scientists to be able to interpret these models. As such, we aim to evaluate and compare different atomic embedding schemes in a consistent framework.
ElementEmbeddings's main feature, the Embedding class is accessible by importing the class.
The latest stable release can be installed via pip using:
pip install ElementEmbeddingsAlternatively, ElementEmbeddings is available via conda through the conda-forge channel on Anaconda Cloud:
conda install -c conda-forge elementembeddingsFor installing the development or documentation dependencies via pip:
pip install "ElementEmbeddings[dev]"
pip install "ElementEmbeddings[docs]"For development, you can clone the repository and install the package in editable mode. To clone the repository and make a local installation, run the following commands:
git clone https://github.com/WMD-group/ElementEmbeddings.git
cd ElementEmbeddings
pip install -e ".[docs,dev]"With -e pip will create links to the source folder so that changes to the code will be immediately reflected on the PATH.
For simple usage, you can instantiate an Embedding object using one of the embeddings in the data directory. For this example, let's use the magpie elemental representation.
# Import the class
>>> from elementembeddings.core import Embedding
# Load the magpie data
>>> magpie = Embedding.load_data('magpie')We can access some of the properties of the Embedding class. For example, we can find the dimensions of the elemental representation and the list of elements for which an embedding exists.
# Print out some of the properties of the ElementEmbeddings class
>>> print(f'The magpie representation has embeddings of dimension {magpie.dim}')
>>> print(f'The magpie representation contains these elements: \n {magpie.element_list}') # prints out all the elements considered for this representation
>>> print(f'The magpie representation contains these features: \n {magpie.feature_labels}') # Prints out the feature labels of the chosen representation
The magpie representation has embeddings of dimension 22
The magpie representation contains these elements:
['H', 'He', 'Li', 'Be', 'B', 'C', 'N', 'O', 'F', 'Ne', 'Na', 'Mg', 'Al', 'Si', 'P', 'S', 'Cl', 'Ar', 'K', 'Ca', 'Sc', 'Ti', 'V', 'Cr', 'Mn', 'Fe', 'Co', 'Ni', 'Cu', 'Zn', 'Ga', 'Ge', 'As', 'Se', 'Br', 'Kr', 'Rb', 'Sr', 'Y', 'Zr', 'Nb', 'Mo', 'Tc', 'Ru', 'Rh', 'Pd', 'Ag', 'Cd', 'In', 'Sn', 'Sb', 'Te', 'I', 'Xe', 'Cs', 'Ba', 'La', 'Ce', 'Pr', 'Nd', 'Pm', 'Sm', 'Eu', 'Gd', 'Tb', 'Dy', 'Ho', 'Er', 'Tm', 'Yb', 'Lu', 'Hf', 'Ta', 'W', 'Re', 'Os', 'Ir', 'Pt', 'Au', 'Hg', 'Tl', 'Pb', 'Bi', 'Po', 'At', 'Rn', 'Fr', 'Ra', 'Ac', 'Th', 'Pa', 'U', 'Np', 'Pu', 'Am', 'Cm', 'Bk']
The magpie representation contains these features:
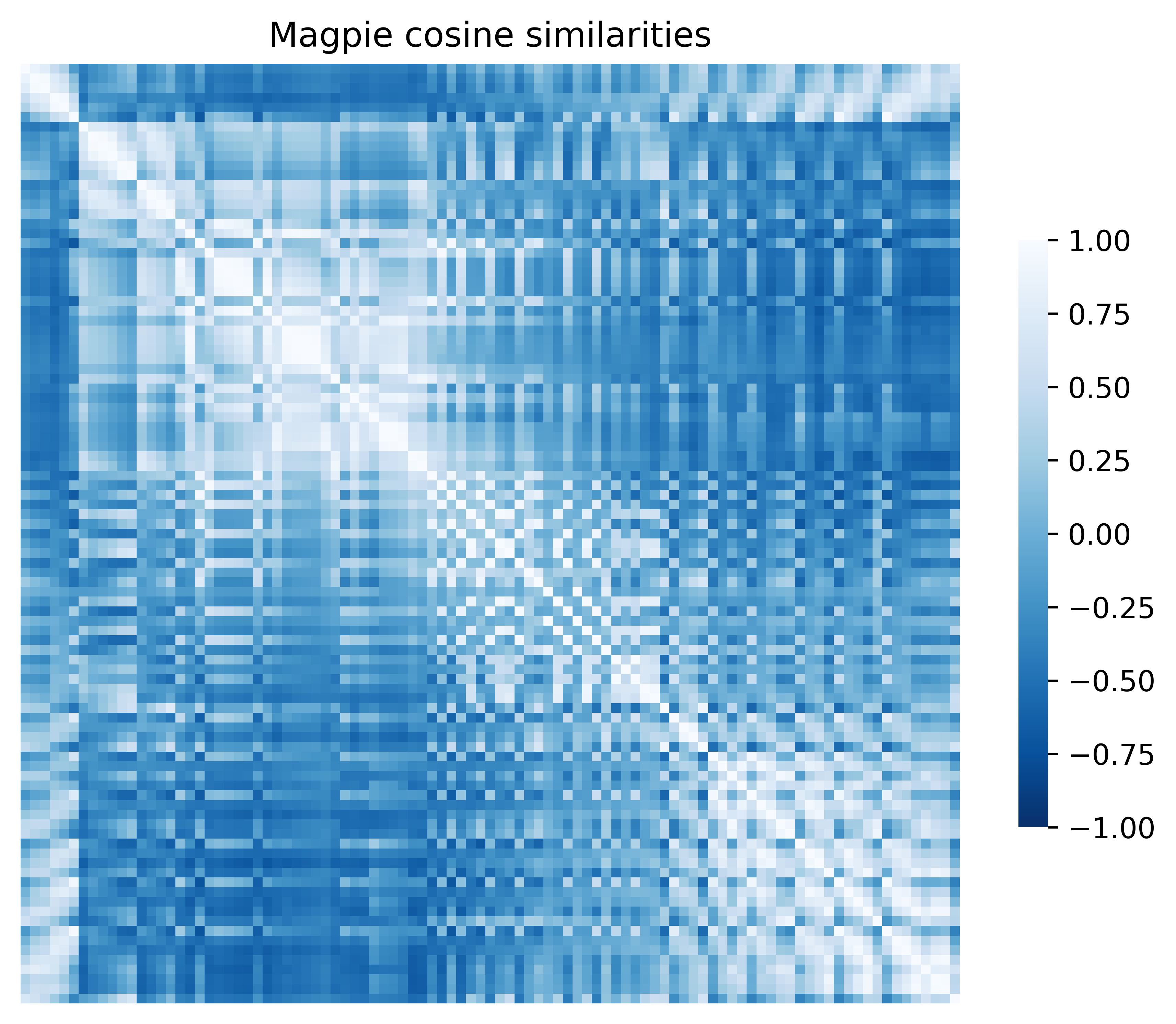
['Number', 'MendeleevNumber', 'AtomicWeight', 'MeltingT', 'Column', 'Row', 'CovalentRadius', 'Electronegativity', 'NsValence', 'NpValence', 'NdValence', 'NfValence', 'NValence', 'NsUnfilled', 'NpUnfilled', 'NdUnfilled', 'NfUnfilled', 'NUnfilled', 'GSvolume_pa', 'GSbandgap', 'GSmagmom', 'SpaceGroupNumber']We can quickly generate heatmaps of distance/similarity measures between the element vectors using heatmap_plotter and plot the representations in two dimensions using the dimension_plotter from the plotter module. Before we do that, we will standardise the embedding using the standardise method available to the Embedding class
from elementembeddings.plotter import heatmap_plotter, dimension_plotter
import matplotlib.pyplot as plt
magpie.standardise(inplace=True) # Standardises the representation
fig, ax = plt.subplots(1, 1, figsize=(6,6))
heatmap_params = {"vmin": -1, "vmax": 1}
heatmap_plotter(embedding=magpie, metric="cosine_similarity",show_axislabels=False,cmap="Blues_r",ax=ax, **heatmap_params)
ax.set_title("Magpie cosine similarities")
fig.tight_layout()
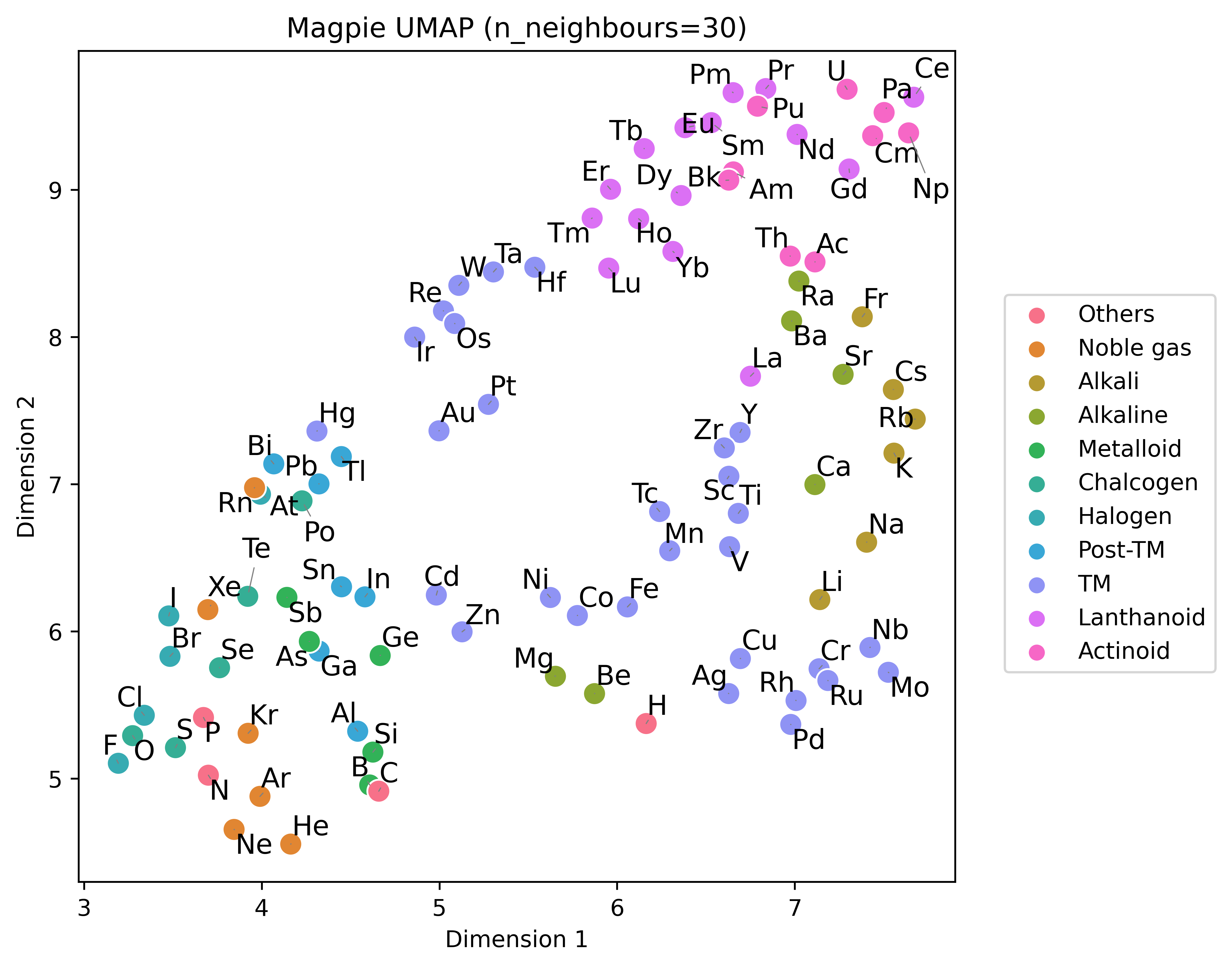
fig.show()fig, ax = plt.subplots(1, 1, figsize=(6,6))
reducer_params={"n_neighbors": 30, "random_state":42}
scatter_params = {"s":100}
dimension_plotter(embedding=magpie, reducer="umap",n_components=2,ax=ax,adjusttext=True,reducer_params=reducer_params, scatter_params=scatter_params)
ax.set_title("Magpie UMAP (n_neighbours=30)")
ax.legend().remove()
handles, labels = ax1.get_legend_handles_labels()
fig.legend(handles, labels, bbox_to_anchor=(1.25, 0.5), loc="center right", ncol=1)
fig.tight_layout()
fig.show()The package can also be used to featurise compositions. Your data could be a list of formula strings or a pandas dataframe of the following format:
| formula |
|---|
| CsPbI3 |
| Fe2O3 |
| NaCl |
| ZnS |
The composition_featuriser function can be used to featurise the data. The compositions can be featurised using different representation schemes and different types of pooling through the embedding and stats arguments respectively.
from elementembeddings.composition import composition_featuriser
df_featurised = composition_featuriser(df, embedding="magpie", stats=["mean","sum"])
df_featurised| formula | mean_Number | mean_MendeleevNumber | mean_AtomicWeight | mean_MeltingT | mean_Column | mean_Row | mean_CovalentRadius | mean_Electronegativity | mean_NsValence | mean_NpValence | mean_NdValence | mean_NfValence | mean_NValence | mean_NsUnfilled | mean_NpUnfilled | mean_NdUnfilled | mean_NfUnfilled | mean_NUnfilled | mean_GSvolume_pa | mean_GSbandgap | mean_GSmagmom | mean_SpaceGroupNumber | sum_Number | sum_MendeleevNumber | sum_AtomicWeight | sum_MeltingT | sum_Column | sum_Row | sum_CovalentRadius | sum_Electronegativity | sum_NsValence | sum_NpValence | sum_NdValence | sum_NfValence | sum_NValence | sum_NsUnfilled | sum_NpUnfilled | sum_NdUnfilled | sum_NfUnfilled | sum_NUnfilled | sum_GSvolume_pa | sum_GSbandgap | sum_GSmagmom | sum_SpaceGroupNumber |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CsPbI3 | 59.2 | 74.8 | 144.16377238 | 412.55 | 13.2 | 5.4 | 161.39999999999998 | 2.22 | 1.8 | 3.4 | 8.0 | 2.8000000000000003 | 16.0 | 0.2 | 1.4 | 0.0 | 0.0 | 1.6 | 54.584 | 0.6372 | 0.0 | 129.20000000000002 | 296.0 | 374.0 | 720.8188619 | 2062.75 | 66.0 | 27.0 | 807.0 | 11.100000000000001 | 9.0 | 17.0 | 40.0 | 14.0 | 80.0 | 1.0 | 7.0 | 0.0 | 0.0 | 8.0 | 272.92 | 3.186 | 0.0 | 646.0 |
| Fe2O3 | 15.2 | 74.19999999999999 | 31.937640000000002 | 757.2800000000001 | 12.8 | 2.8 | 92.4 | 2.7960000000000003 | 2.0 | 2.4 | 2.4000000000000004 | 0.0 | 6.8 | 0.0 | 1.2 | 1.6 | 0.0 | 2.8 | 9.755 | 0.0 | 0.8442651200000001 | 98.80000000000001 | 76.0 | 371.0 | 159.6882 | 3786.4 | 64.0 | 14.0 | 462.0 | 13.98 | 10.0 | 12.0 | 12.0 | 0.0 | 34.0 | 0.0 | 6.0 | 8.0 | 0.0 | 14.0 | 48.775000000000006 | 0.0 | 4.2213256 | 494.0 |
| NaCl | 14.0 | 48.0 | 29.221384640000004 | 271.235 | 9.0 | 3.0 | 134.0 | 2.045 | 1.5 | 2.5 | 0.0 | 0.0 | 4.0 | 0.5 | 0.5 | 0.0 | 0.0 | 1.0 | 26.87041666665 | 1.2465 | 0.0 | 146.5 | 28.0 | 96.0 | 58.44276928000001 | 542.47 | 18.0 | 6.0 | 268.0 | 4.09 | 3.0 | 5.0 | 0.0 | 0.0 | 8.0 | 1.0 | 1.0 | 0.0 | 0.0 | 2.0 | 53.7408333333 | 2.493 | 0.0 | 293.0 |
| ZnS | 23.0 | 78.5 | 48.7225 | 540.52 | 14.0 | 3.5 | 113.5 | 2.115 | 2.0 | 2.0 | 5.0 | 0.0 | 9.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 19.8734375 | 1.101 | 0.0 | 132.0 | 46.0 | 157.0 | 97.445 | 1081.04 | 28.0 | 7.0 | 227.0 | 4.23 | 4.0 | 4.0 | 10.0 | 0.0 | 18.0 | 0.0 | 2.0 | 0.0 | 0.0 | 2.0 | 39.746875 | 2.202 | 0.0 | 264.0 |
The returned dataframe contains the mean-pooled and sum-pooled features of the magpie representation for the four formulas.
Please use the issue tracker to report bugs and any feature requests. Hopefully, most questions should be solvable through the docs. For any other queries related to the project, please contact Anthony Onwuli by e-mail: anthony.onwuli16@imperial.ac.uk.
We welcome new contributions to this project. See the contributing guide for detailed instructions on how to contribute to our project.
The steps required to add a new representation scheme are:
- Add data file to data/element_representations.
- Edit docstring table in core.py.
- Edit utils/config.py to include the representation in
DEFAULT_ELEMENT_EMBEDDINGSandCITATIONS. - Update the documentation reference.md and README.md.
- Anthony Onwuli (Department of Materials, Imperial College London)
A. Onwuli et al, "Ionic species representations for materials informatics"
H. Park et al, "Mapping inorganic crystal chemical space" Faraday Discuss. (2024)