To understand immune activation and evasion mechanisms in cancer, one crucial step is to characterize the composition of immune and stromal cells in the tumor microenvironment (TME). Deconvolution analysis based on bulk transcriptomic data has been used to estimate cell composition in TME. However, these algorithms are sub-optimal for proteomic data, which has hindered research in the rapidly growing field of proteogenomics. Moreover, with the increasing prevalence of multi-omics studies, there is an opportunity to enhance deconvolution analysis by utilizing paired proteomic and transcriptomic profiles of the same tissue samples. To bridge these gaps, we propose BayesDeBulk, a new method for estimating the immune/stromal cell composition based on bulk proteomic and gene expression data. BayesDeBulk utilizes the information of known cell-type-specific markers without requiring their absolute abundance levels as prior knowledge.
For more information, please visit or cite the related preprint:
- library(devtools)
- install_github("WangLab-MSSM/BayesDeBulk/BayesDeBulk")
- Requires R >= 3.6
The following command will perform tumor deconvolution with an input signature matrix for combined multi-omic data, including a protein abundance file and RNA expression file.
cd R
Rscript main.R --multiomic=TRUE --abundanceFile='../test_data/proteo_dummy.tsv' --expressionFile='../test_data/RNA_dummy.tsv' --signatureMatrix='../test_data/LM22_combined_cell_types.tsv' --rowMeansImputation=TRUEThe code and test data are available as a Docker image tagged cptacdream/bayesdebulk.
| flag | type | default | description |
|---|---|---|---|
| multiomic | boolean | FALSE | indicates whether to compute tumor deconvolution with both RNA expression and protein abundance |
| abundanceFile | filepath | path to tab-separated protein abundance table file, where rows are gene symbols and columns are samples | |
| expressionFile | filepath | path to tab-separated RNA expression table file, where rows are gene symbols and columns are samples | |
| signatureMatrix | filepath | path to signature matrix table file, where rows are gene symbols and columns are cell types | |
| rowMeansImputation | boolean | TRUE | indicates whether to perform row means imputation for NA values in -omics files |
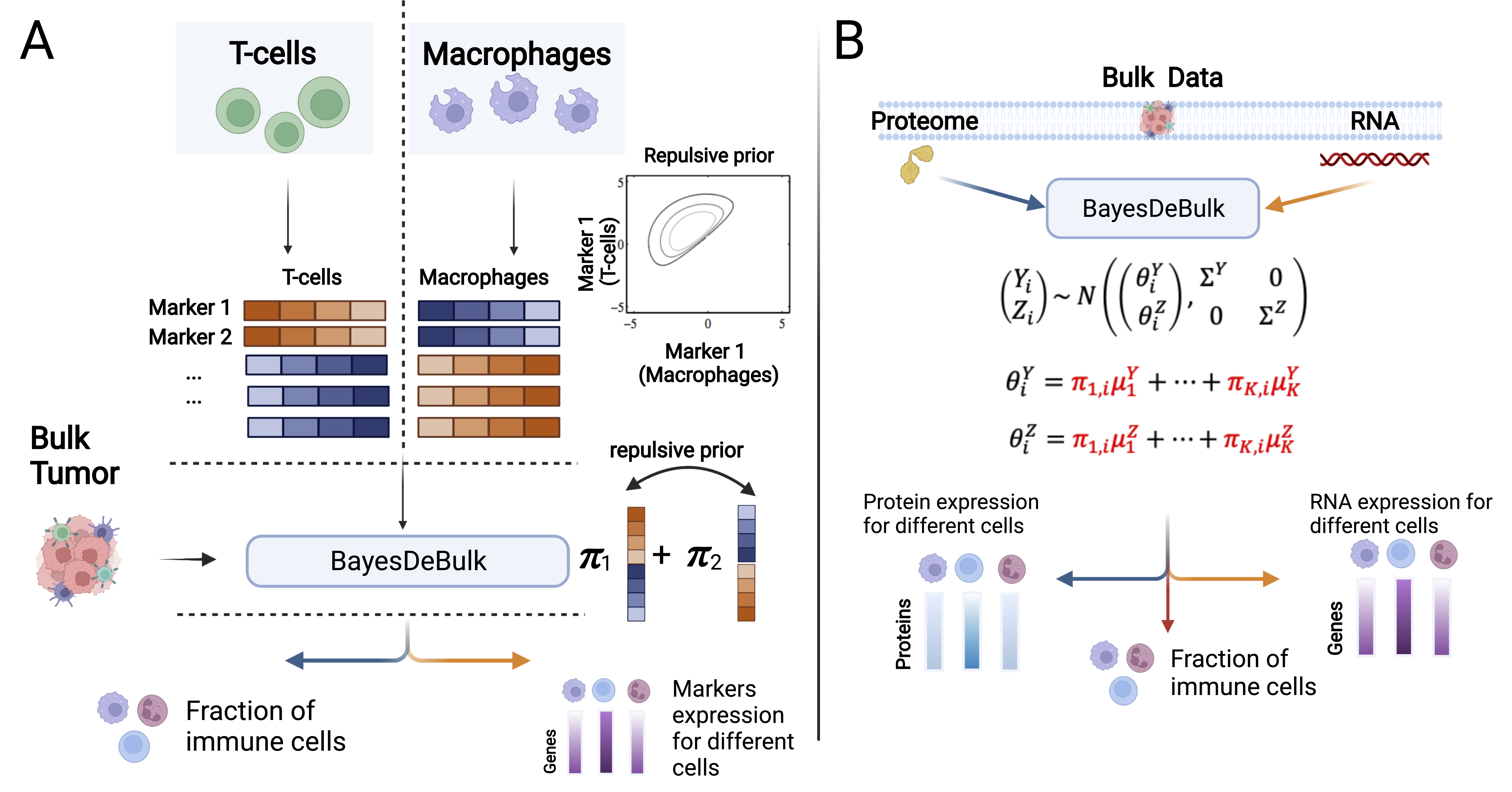
Algorithm Schematic. (A) Bulk data is modeled as a linear combination of marker expression in different cell types. Given a list of markers expressed in each cell type, a Repulsive prior is placed on the mean of marker expression in different cell types to ensure that cell type specific markers are upregulated in a particular component. (B) Multi-omic framework to estimate cell type fractions integrating proteomic and RNAseq data. Given a list of cell-type specific markers, the algorithm returns the estimated protein/RNA expression for different cell types and cell-type fractions for different samples.