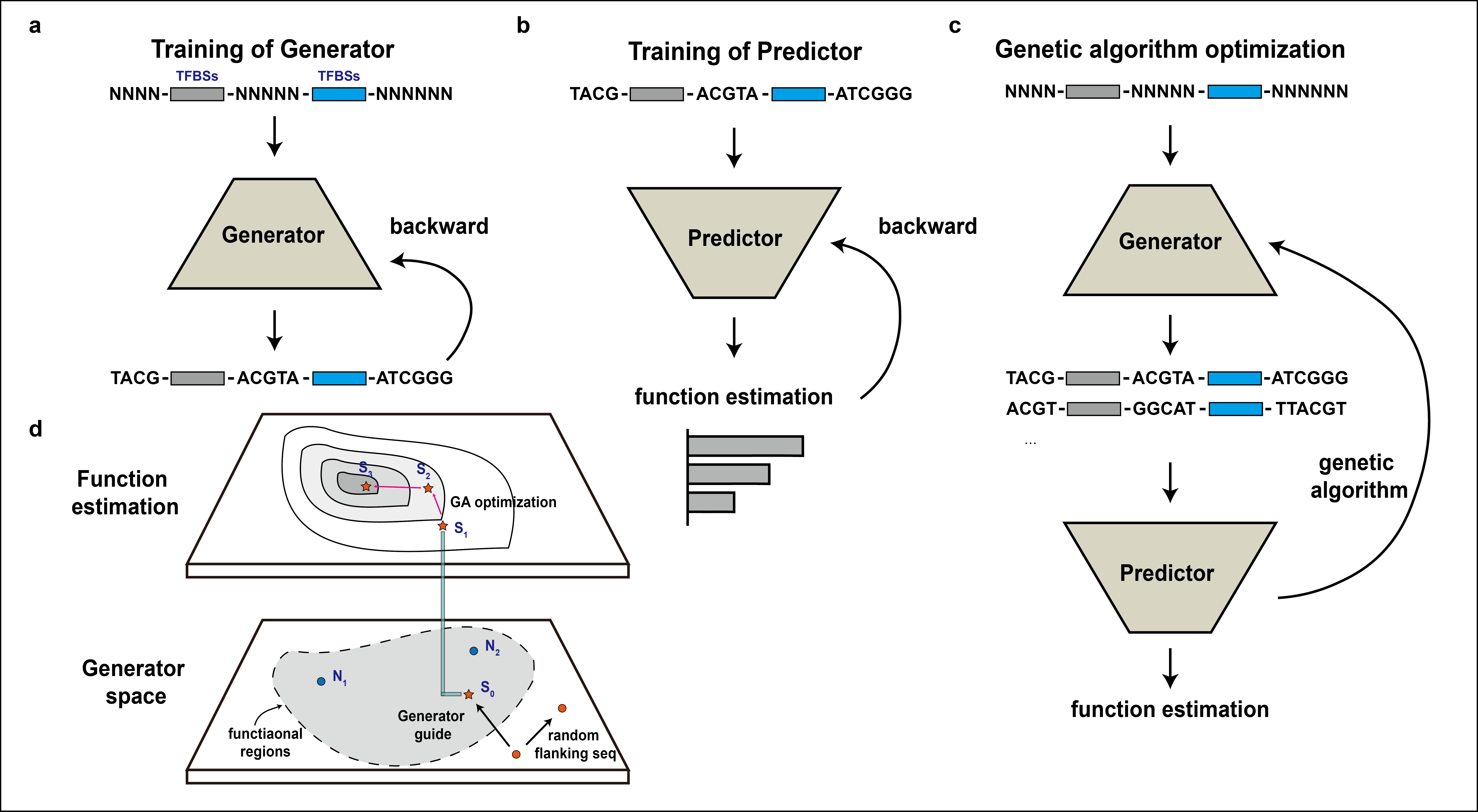
The code for official implementation of "Deep flanking sequences engineering for efficient promoter design" This codebase provides:
- The conditional generative adversarial networks (GANs) that generate promoter sequences when giving conditional motif sequences. (the folder "Generator")
- The predictor that could evaluate promoter activities based on densenet-lstm model. (the folder "Predictor")
- The optimizer that could design promoter sequences with high activities when giving conditional motif sequences. (the folder "Optimizer")
We introduced an AI-aided promoter design framework, DeepSEED, that employs both experts’ knowledge and deep learning together to efficiently design synthetic promoters of diverse desirable functions. DeepSEED incorporates the user-defined cis-regulatory sequences as ‘seed’ and generates flanking sequences that match the ‘seed’. We showed that DeepSEED could automatically capture a variety of weak patterns like k-mer frequencies and DNA shape features from active promoters in the training set, and efficiently optimize the flanking sequences in synthetic promoters to better match these properties.
We validated the effectiveness of this framework for diverse synthetic promoter design tasks in both prokaryotic and eukaryotic cells. DeepSEED successfully designed constitutive, IPTG-inducible, and doxycycline(Dox)-inducible promoters with significant performance improvements, suggesting DeepSEED as an efficient AI-aided flanking sequence optimization approach for promoter design that may greatly benefit synthetic biology applications.
Env Requirements:
- MAC OS, Linux or Windows.
- Python 3.7+.
- PyTorch 1.7.1
- CUDA 11.2 if you need train deep learning model with gpu.
Steps of using DeepSEED:
-
Install Python ref to Download Python
-
Install DeepSEED in virtualenv to keep your environment clean:
pip install virtualenv # or pip install -i https://pypi.tuna.tsinghua.edu.cn/simple virtualenv virtualenv --python=python3 DeepSEED cd DeepSEED source ./bin/activateOptional: After use, shutdown virtual environment with
deactivatepip install virtualenv # or pip install -i https://pypi.tuna.tsinghua.edu.cn/simple virtualenv virtualenv --python=python3 DeepSEED cd DeepSEED .\Scripts\activate.batOptional: Shutdown virtual environment with
.\Scripts\deactivate.bat -
Install Git, this step is optional if you does not install DeepSEED by git clone. Clone the source codes with git.
git clone https://github.com/WangLabTHU/deepseed.git -
or, download the source codes and extract files and put it in the virtual environment directory you defined.
-
after 2/3, the directory of DeepSEED should have the following structure:
DeepSEED deepseed |------- | |-------data |... |-------Generator |... |-------Predictor |... |-------Optimizer |... |------- |... |... -
After the extraction, download all dependencies with the following commend.
cd deepseed (or cd deepseed-main) pip install -r requirements.txtTo boost download speeds, you could setup pip mirror such as
pip install -i https://pypi.tuna.tsinghua.edu.cn/simple -r requirements.txt # or pip install -i https://mirrors.aliyun.com/pypi/simple -r requirements.txtIt is recommended to use the official website to install pytorch.
# CPU version pip install torch==1.7.1+cpu torchvision==0.8.2+cpu -f https://download.pytorch.org/whl/torch_stable.html # GPU version pip install torch==1.7.1 torchvision==0.8.2 -f https://download.pytorch.org/whl/torch_stable.html
The typical install time on a desktop computer depends on your download speed. Suppose your computer has 1MB/s when downloading python package, it will take about 1 hour to install all the necessary packages.
- Run deepseed with python and enjoy it with following steps:
We take design of 3-lacO IPTG-inducible promoters in E. coli as an example, to illustrate how to train the DeepSEED model and design the promoter sequences
```
cd Generator
python cGAN_training.py
```
```
cd ../Predictor
python predictor_training.py
```
```
cd ../Optimizer
python deepseed_optimizer.py
```
```
vi ./results/ecoli_3_laco.txt
```
Please consider citing our paper in your publications if the project helps your research. BibTeX reference is as follows.
@article{,
title={Deep flanking sequences engineering for efficient promoter design},
author={Pengcheng Zhang, Haochen Wang, Hanwen Xu, Lei Wei, Zhirui Hu, Xiaowo Wang
},
journal={biorkiv},
year={2023}
}
For academic use, this project is licensed under the MIT License - see the LICENSE file for details For commercial use, please contact the authors.
[1] Isola, Phillip, et al. "Image-to-image translation with conditional adversarial networks." Proceedings of the IEEE conference on computer vision and pattern recognition. 2017.
Parts of the code have been modified from the Pix2pix, which could be found at https://github.com/phillipi/pix2pix.