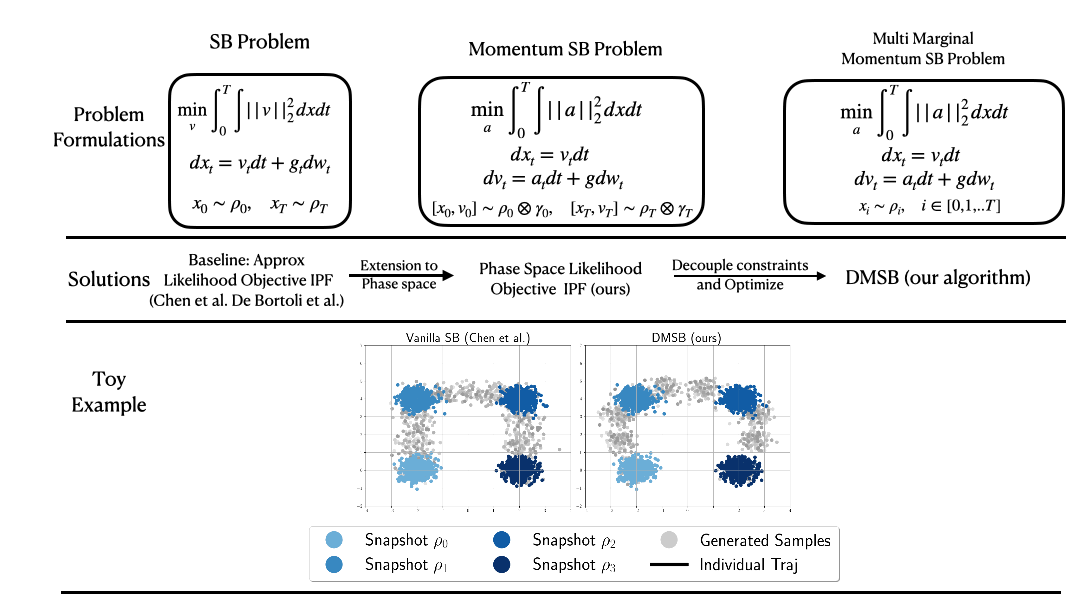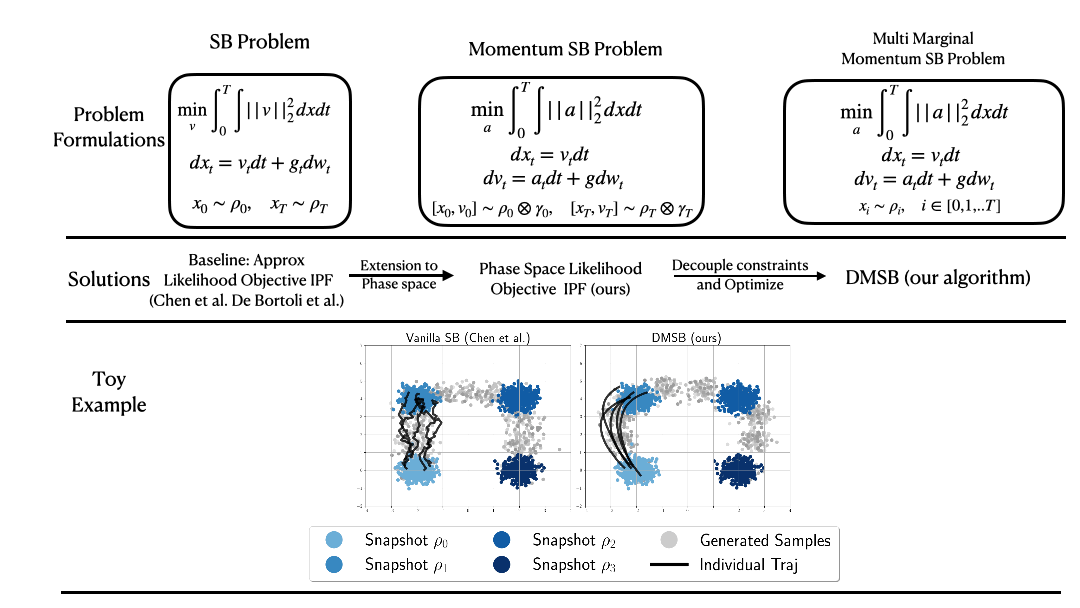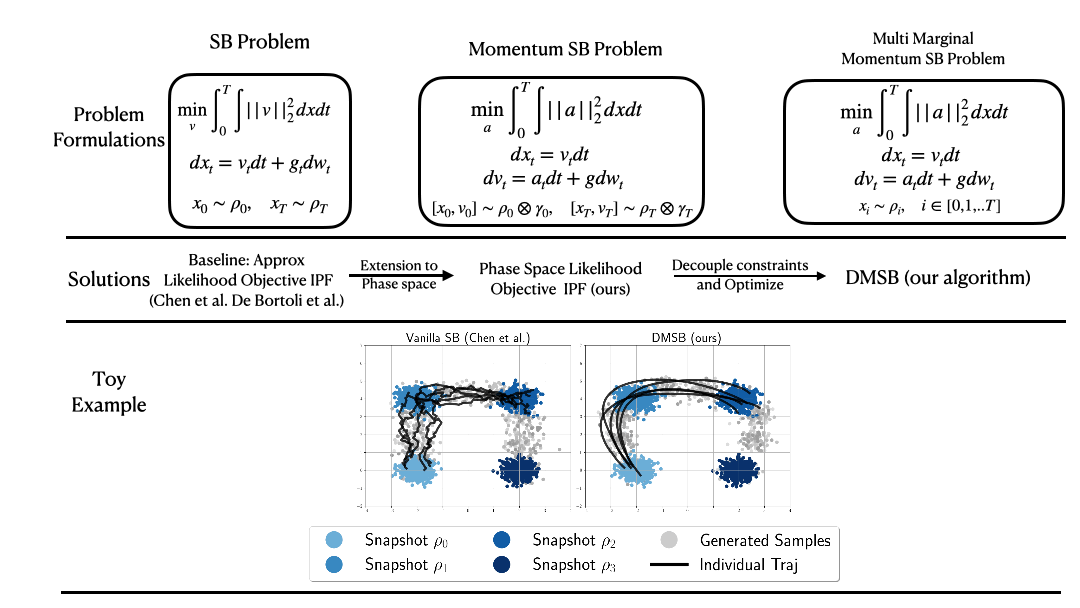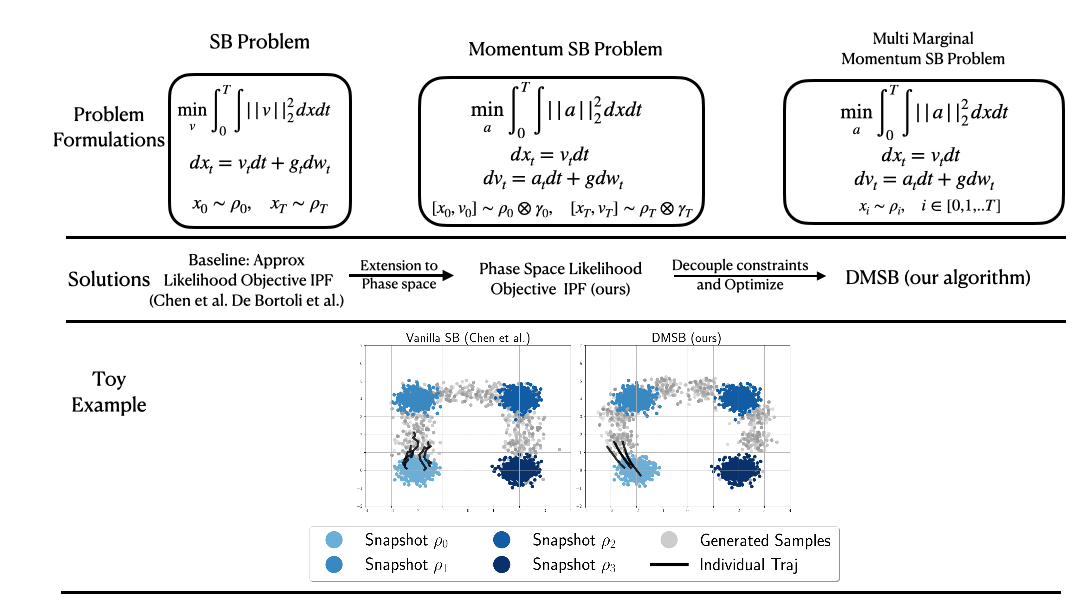
(NeurIPS2023) DMSB: Deep Momentum Multi-Marginal Schrödinger Bridge [LINK]
Official PyTorch implementation of the paper "Deep Momentum Multi-Marginal Schrödinger Bridge (DMSB)" which introduces a new class of trajectory inference models that extend SB models to momentum dynamcis and multi-marginal case.
Tasks (--problem-name) |
Results |
|---|---|
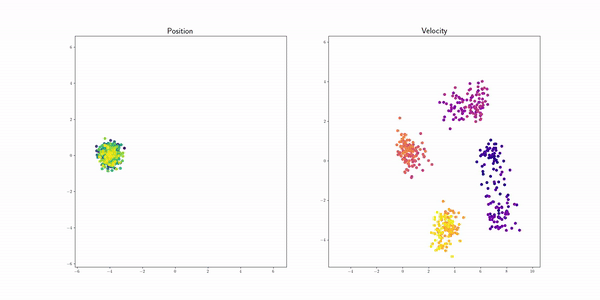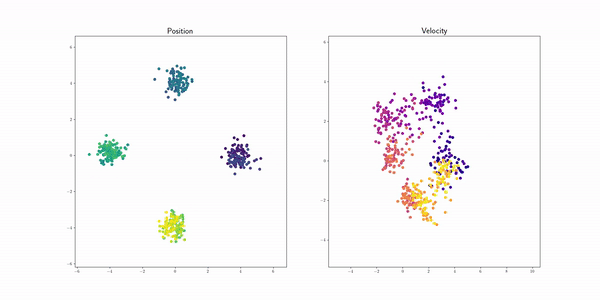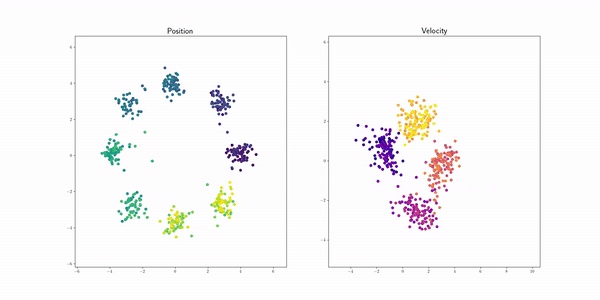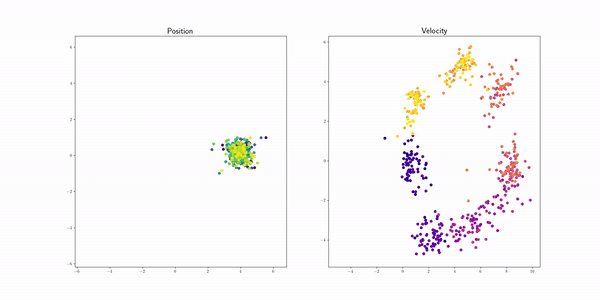
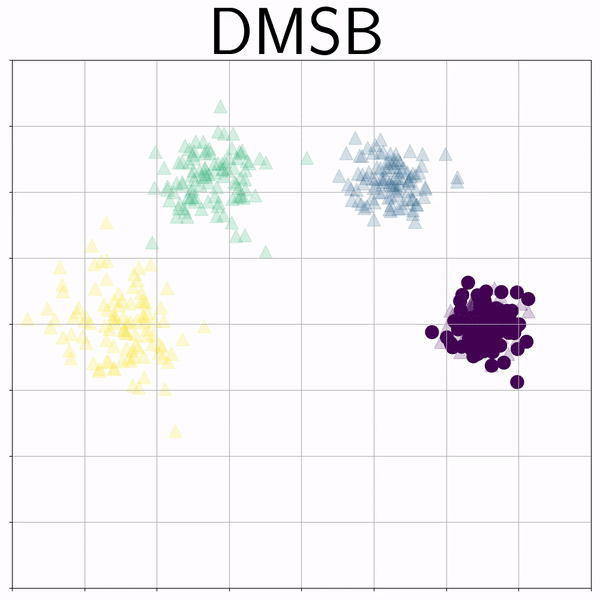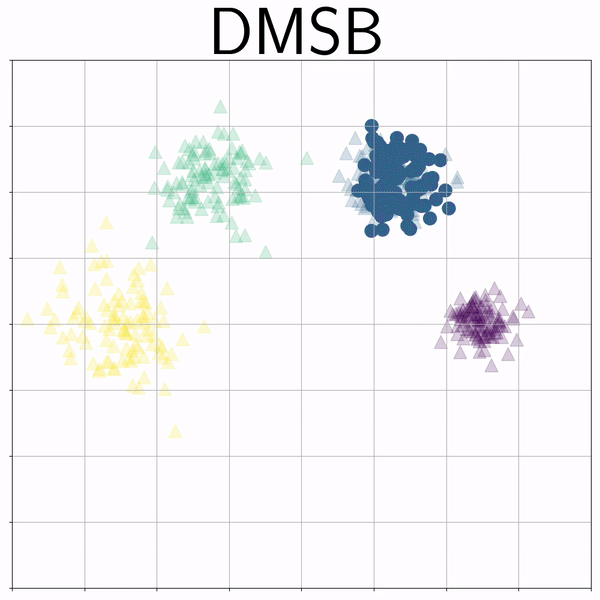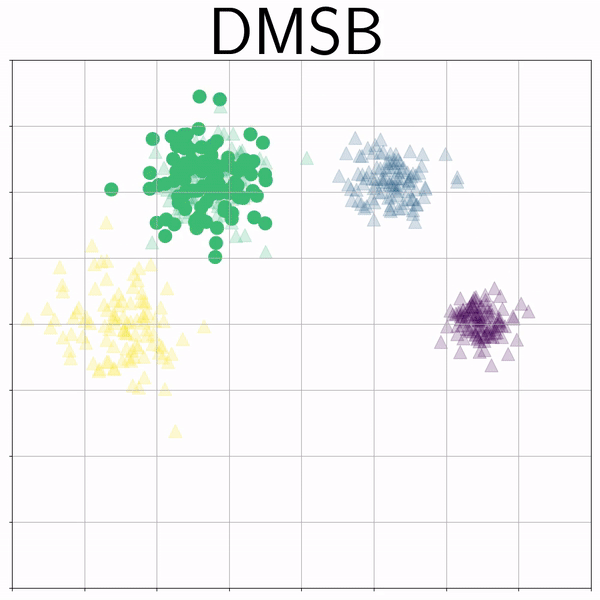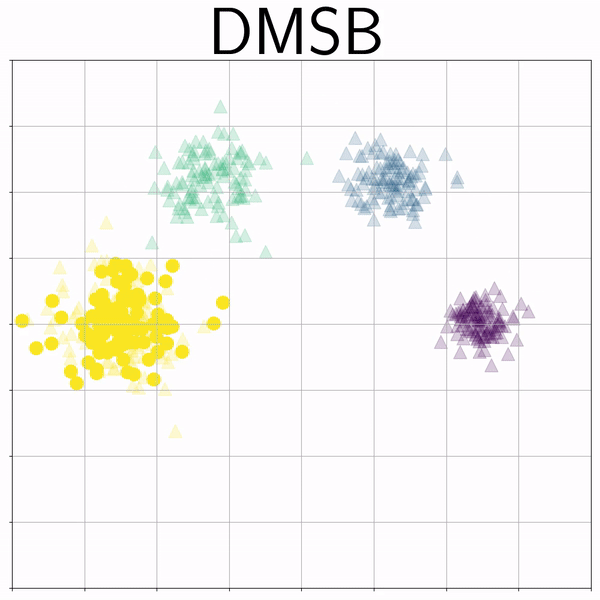
Mixture Gaussians (gmm) |
|
Semicircle (semicircle) |
|
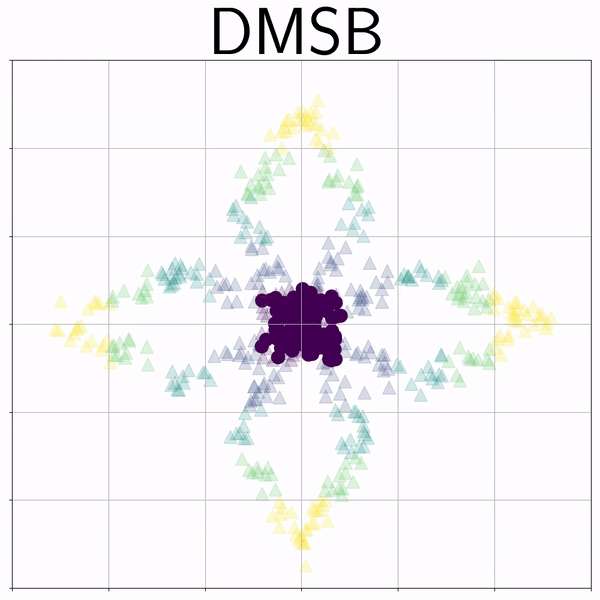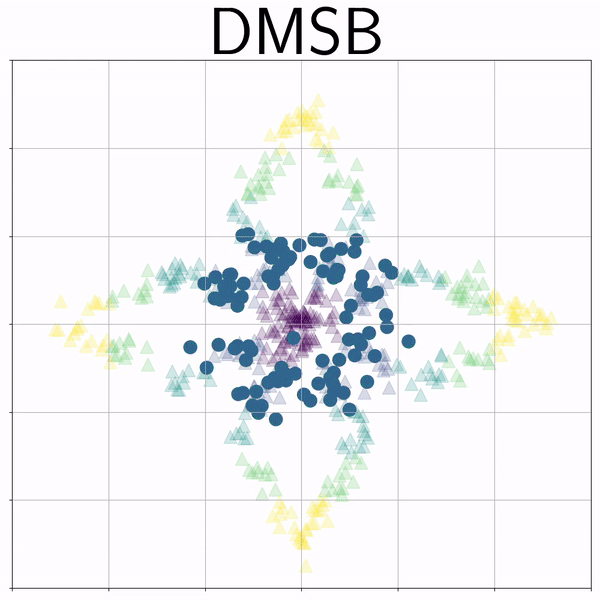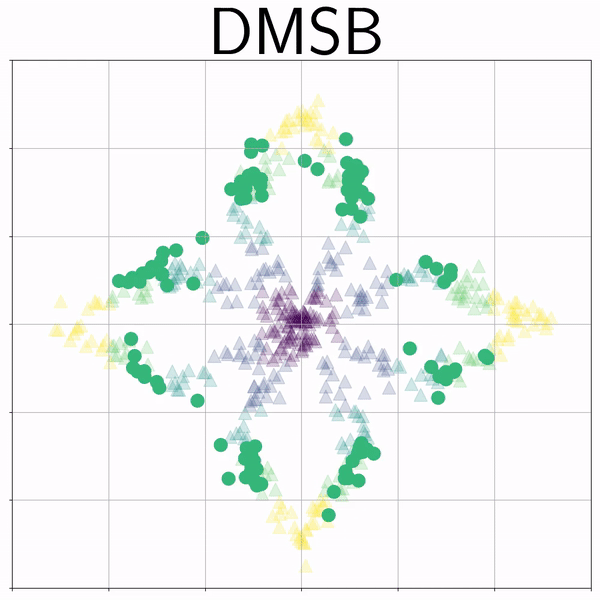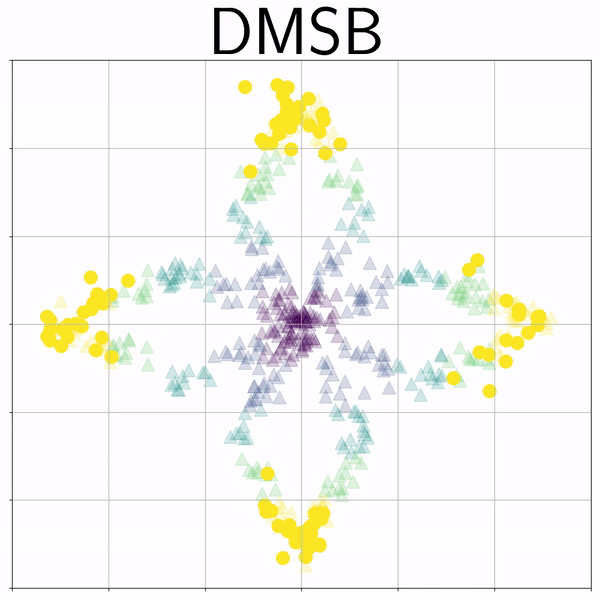
Petal (Petal) |
|
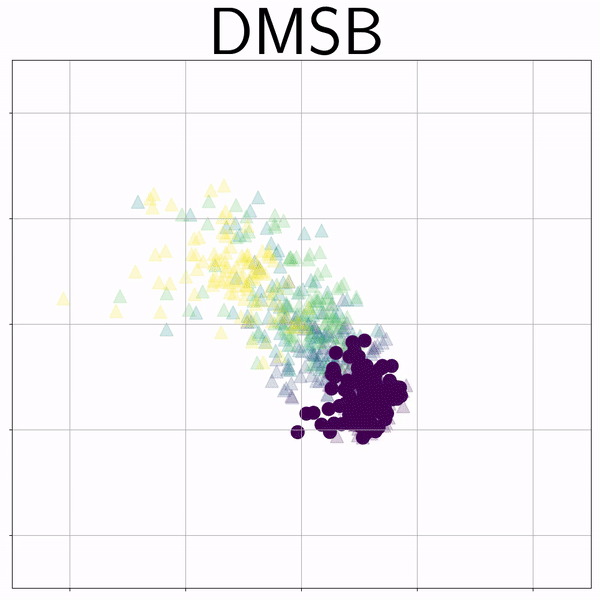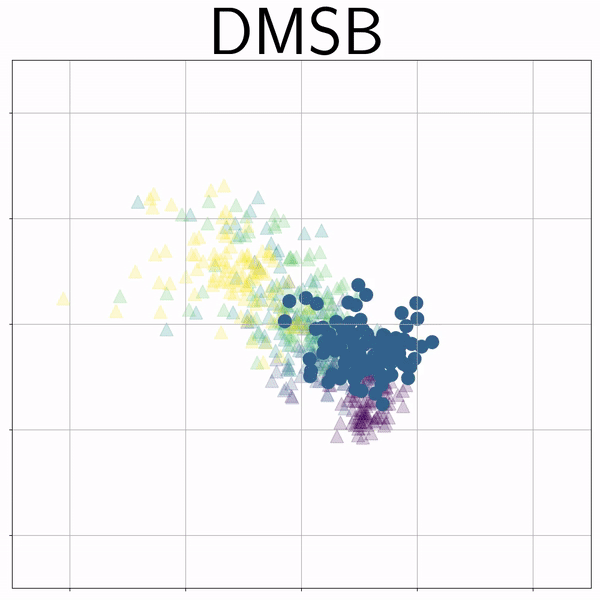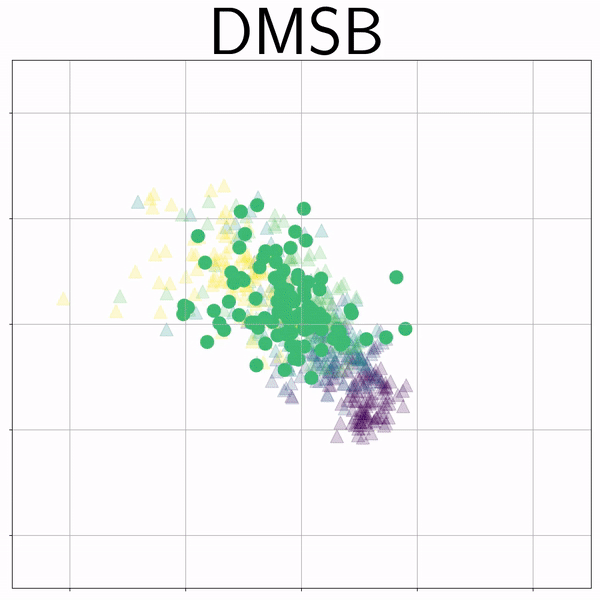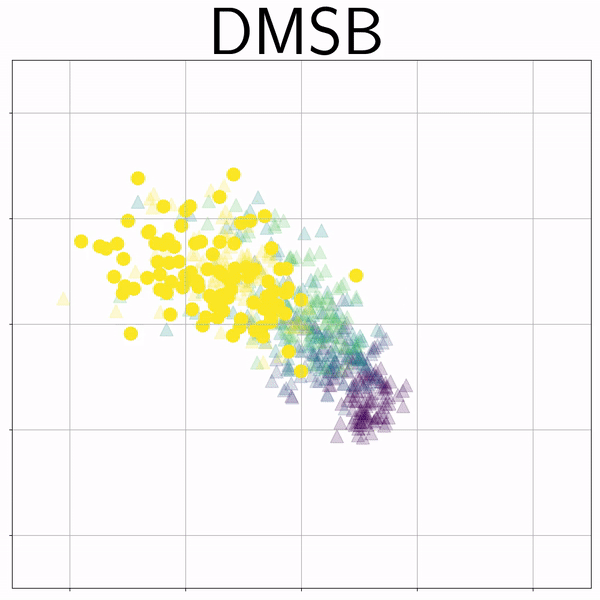
100-Dim Single Cell RNA sequence (RNAsc) |
|
| If you find this library useful, please cite ⬇️ |
@article{chen2023deep,
title={Deep Momentum Multi-Marginal Schr$\backslash$" odinger Bridge},
author={Chen, Tianrong and Liu, Guan-Horng and Tao, Molei and Theodorou, Evangelos A},
journal={arXiv preprint arXiv:2303.01751},
year={2023}
}
(Environment may have conflict with cuda version... I am currently fixing it... but it should work for most of cuda...)This code is developed with Python3. PyTorch >=1.7 (we recommend 1.8.1). First, install the dependencies with Anaconda and activate the environment DMSB with
conda env create --file requirements.yaml python=3.8
conda activate DMSBDownload the RNA-seq daaset from this repo, and put it under ./data/RNAsc/ProcessedData/.
We provide the checkpoint and the code for training from scratch for all the dataset reported in the paper.
python main.py --problem-name gmm --dir reproduce/gmm --log-tb --gpu 1Memo: The results in the paper sould be reproduced by around 6 stage of Bregman Iteration.
python main.py --problem-name petal --dir reproduce/petal --log-tbMemo: The results in the paper sould be reproduced by around 17 stage of Bregman Iteration.
python main.py --problem-name RNAsc --dir reproduce/RNA --log-tb --num-itr 2000
python main.py --problem-name RNAsc --dir reproduce/RNA-loo1 --log-tb --use-amp --num-itr 2000 --LOO 1
python main.py --problem-name RNAsc --dir reproduce/RNA-loo2 --log-tb --use-amp --num-itr 2000 --LOO 2
python main.py --problem-name RNAsc --dir reproduce/RNA-loo3 --log-tb --use-amp --num-itr 2000 --LOO 3The visualization results are saved in the folder /results.
The numerical value are saved in the tensorboard and event file are saved the folder /runs,
The checkpoints are saved in the folder /checkpoint, and you can reload the checkpoint by:
python main.py --problem-name [problem-name] --dir [your/dir/name/for/current/run] --log-tb --load [dir/to/checkpoints/]The numerical results for all metrics will be displayed in the terminal as well.