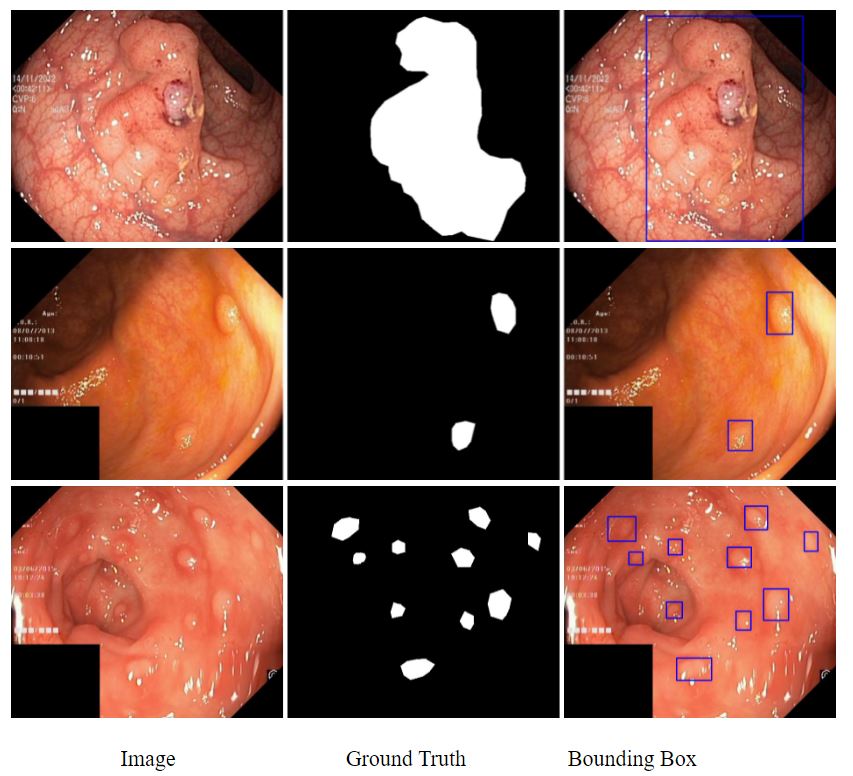
A segmentation model of detecting polyps in colonoscopy developed by Clara Train SDK v3.1.
https://ngc.nvidia.com/catalog/containers/nvidia:clara-train-sdk
https://docs.nvidia.com/clara/tlt-mi/
A brief introduction for this project:
https://docs.google.com/presentation/d/16lPDuL_5Fk6vx14dmSBLxgIIIEO4zSMs5hLENdoX8TQ/edit?usp=sharing
The dataset we used is Kvasir-SEG. The Kvasir-SEG dataset (size 46.2 MB) contains 1000 polyp images and their corresponding ground truth from the Kvasir Dataset v2. The resolution of the images contained in Kvasir-SEG varies from 332x487 to 1920x1072 pixels. https://datasets.simula.no/kvasir-seg/
Debesh Jha, Pia H. Smedsrud, Michael A. Riegler, Pål Halvorsen, Dag Johansen, Thomas de Lange, and Håvard D. Johansen, Kvasir-SEG: A Segmented Polyp Dataset, In Proceedings of the ternational conference on Multimedia Modeling, Republic of Korea, 2020.
For constructing the config of datalist easily, we can:
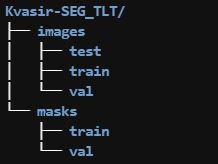
- Prepare the folder structure as below,
- Split the dataset into training, validation and testing set,
- And convert the image format to PNG.
Just follow the notebook to prepare the data.
In Clara Train, the Medical Model ARchive, MMAR, defines a standard structure for organizing all artifacts produced during the model development life cycle. We can define the pre-transformation, hyperparameters, loss function, model, metrics, data root in the configs and simply run the train.sh to train a model. Follow the commands below in Clara Train SDK v3.1 container to start the training:
cd Colonoscopy_MMAR_v1/commands/
chmod +x train.sh
./train.sh