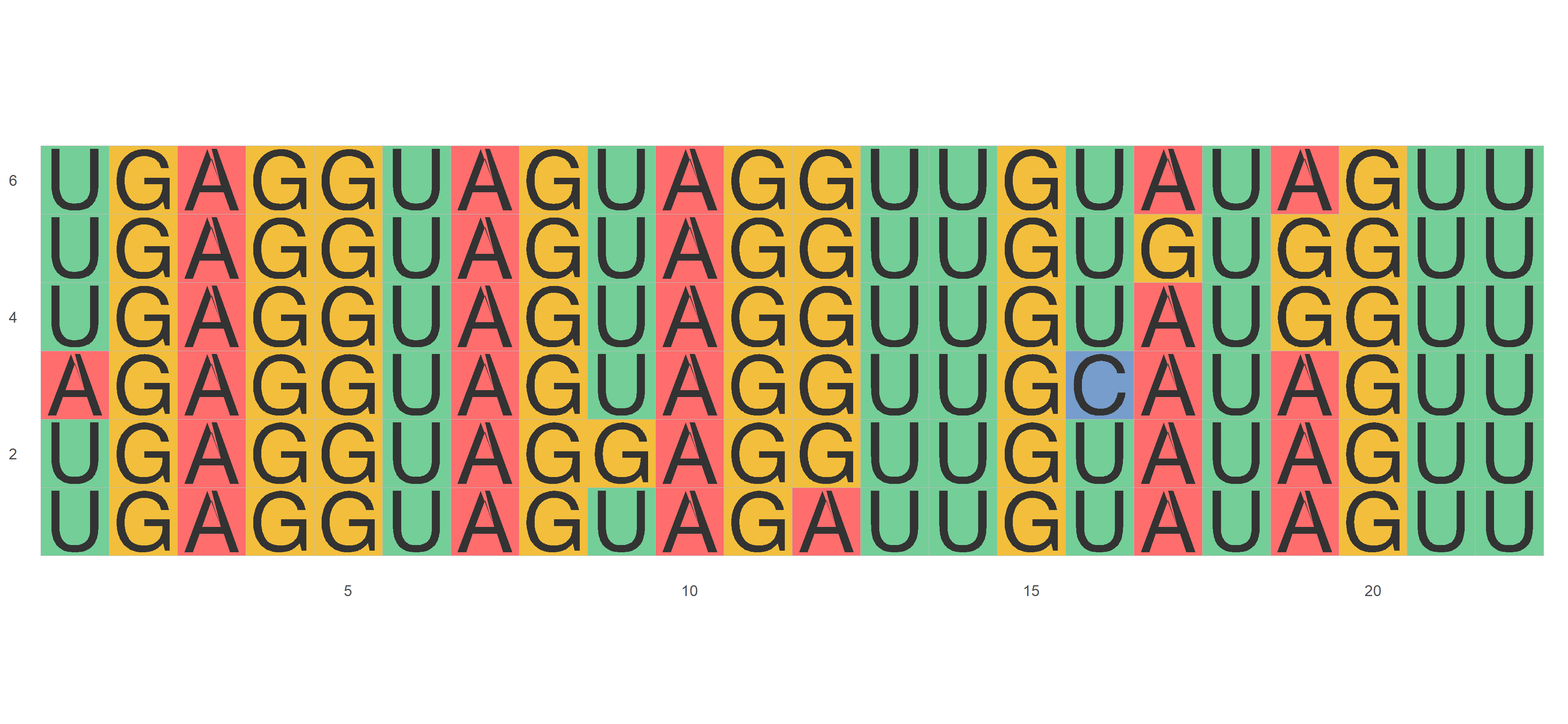ggmsa supports visualizing multiple sequence alignment of nucleotide
sequences and protein sequences using ggplot2. It supports a number of
colour schemes, including Chemistry, Clustal, Shapely, Taylor and Zappo.
The development version from github:
if (!requireNamespace("devtools", quietly=TRUE))
install.packages("devtools")
devtools::install_github("YuLab-SMU/ggmsa")Plot multiple sequence alignment.
library(ggmsa)
library(ggplot2)
protein_sequences <- system.file("extdata", "sample.fasta", package = "ggmsa")
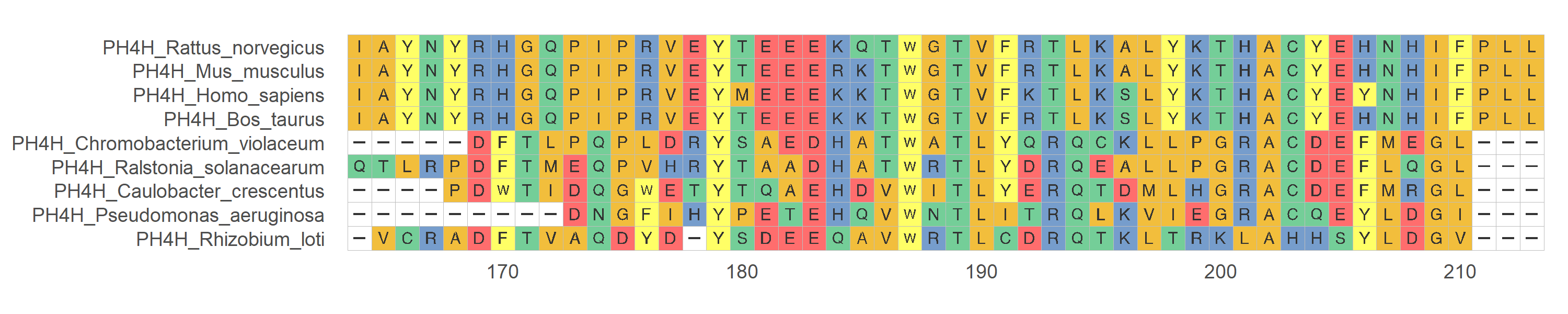
ggmsa(protein_sequences, 164, 213, char_width = 0.5, color = "Chemistry_AA", seq_name = T)nt_sequences <- system.file("extdata", "LeaderRepeat_All.fa", package = "ggmsa")
ggmsa(nt_sequences,font = NULL, color = "Chemistry_NT")miRNA_sequences <- system.file("extdata", "seedSample.fa", package = "ggmsa")
ggmsa(miRNA_sequences, color = "Chemistry_NT")library(Biostrings)
x <- readAAStringSet(protein_sequences)
d <- as.dist(stringDist(x, method = "hamming")/width(x)[1])
library(ape)
tree <- bionj(d)
library(ggtree)
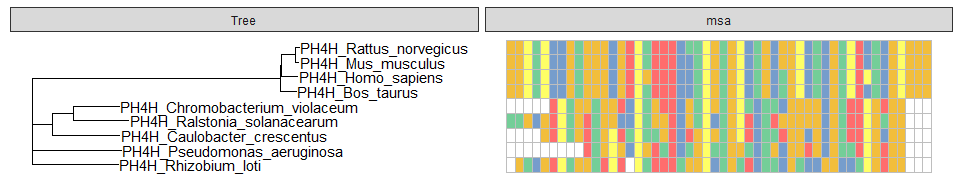
p <- ggtree(tree) + geom_tiplab()
data = tidy_msa(x, 164, 213)
p + geom_facet(geom = geom_msa, data = data, panel = 'msa',
font = NULL, color = "Chemistry_AA") +
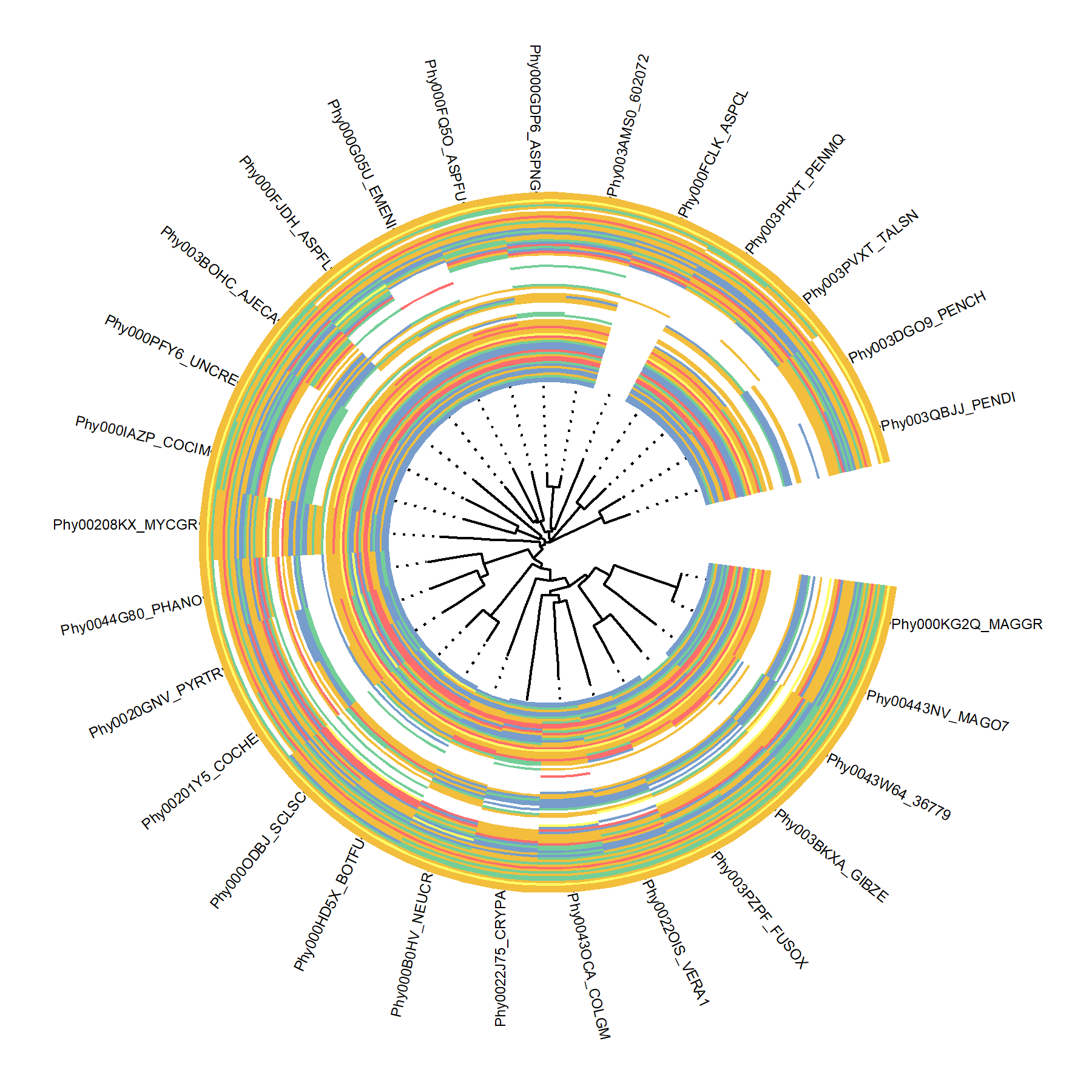
xlim_tree(1)ggmsa also allows MSA graphs to align to the tree with circular, fan, or radial layout by ggtreeExtra(ver >= 1.1.3.991).
library(ggplot2)
library(ggtree)
library(ggtreeExtra)
library(Biostrings)
library(ape)
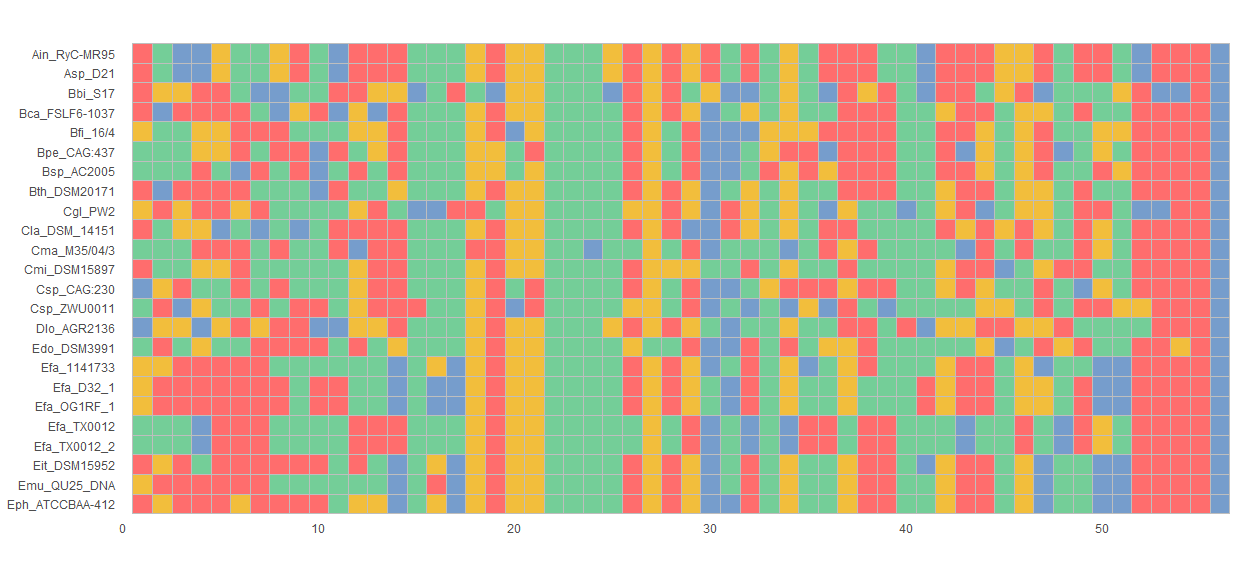
sequences <- system.file("extdata", "sequence-link-tree.fasta", package = "ggmsa")
x <- readAAStringSet(sequences)
d <- as.dist(stringDist(x, method = "hamming")/width(x)[1])
tree <- bionj(d)
data <- tidy_msa(x, 120, 200)
p1 <- ggtree(tree, layout = 'circular') + geom_tiplab(align = TRUE, offset = 0.545, size = 2) + xlim(NA, 1.2)
p1 + geom_fruit(data = data, geom = geom_msa, offset = 0, pwidth = 1.2, font = NULL, border = NA)For more details about the version in CRAN, please refer to the online vignette
Moreover, check out the guides for learning new features with the current development version: