Webb laboratory and Singh laboratory @Brown
Using Brown's HPC (OSCAR)
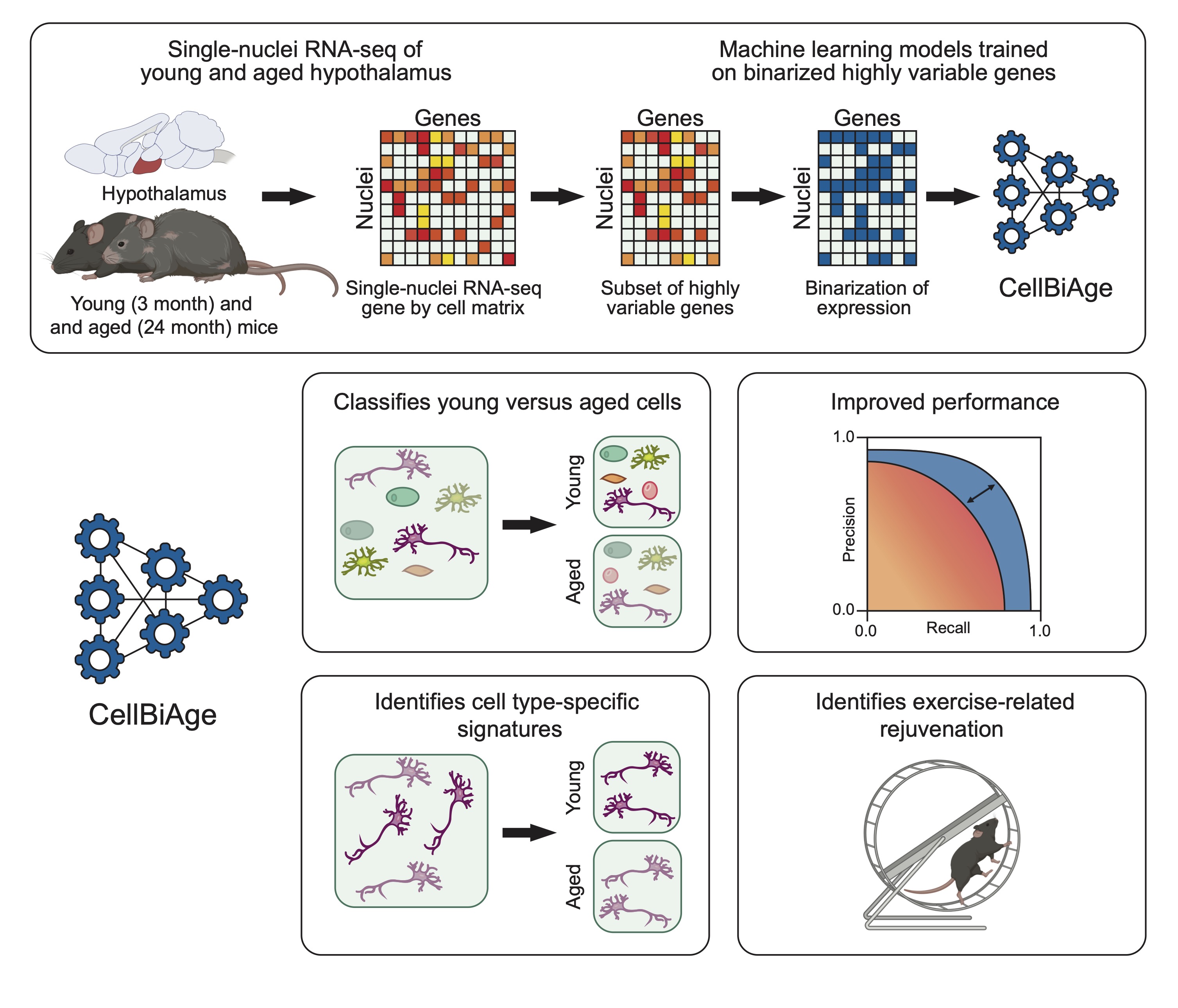
ML/DL applications in predicting cellcular age using mouse brain single cell/nuclei RNA-seq.
- Logistic regression with regularization
- Tree-based models
- Support Vector Machine Classifier (SVC)
- Multilayer Perceptron (MLP)
Other required packages for GPU:
module load cuda/11.7.1
module load cudnn/8.2.0
In terminal: To implement MLP KerasTuner for group-based cross validation:
cd scripts
python3 mlp_kt_4cv_console.py
To implement the best MLP over 10 random seeds:
python3 mlp_rs_console.py
0x: different preprocessing methods1x: hypothalamus all-cell models2x: hypothalamus cell type-specific models3x: SVZ all-cell models4x: SVZ cell type-specific models5x: Bechmarking results6x: Batch integration and misc
4. Datasets
Hajdarovic, K. H., Yu, D., Hassell, L. A., Evans, S. A., Packer, S., Neretti, N., & Webb, A. E. (2022). Single-cell analysis of the aging female mouse hypothalamus. Nature Aging, 2(7), 662-678.
Buckley, M. T., Sun, E. D., George, B. M., Liu, L., Schaum, N., Xu, L., ... & Brunet, A. (2023). Cell-type-specific aging clocks to quantify aging and rejuvenation in neurogenic regions of the brain. Nature Aging, 3(1), 121-137.