Title - Multiscale Topology-enabled Structure-to-Sequence Transformer for Protein-Ligand Interaction Predictions.
Authors - Dong Chen, Jian Liu, and Guo-wei Wei
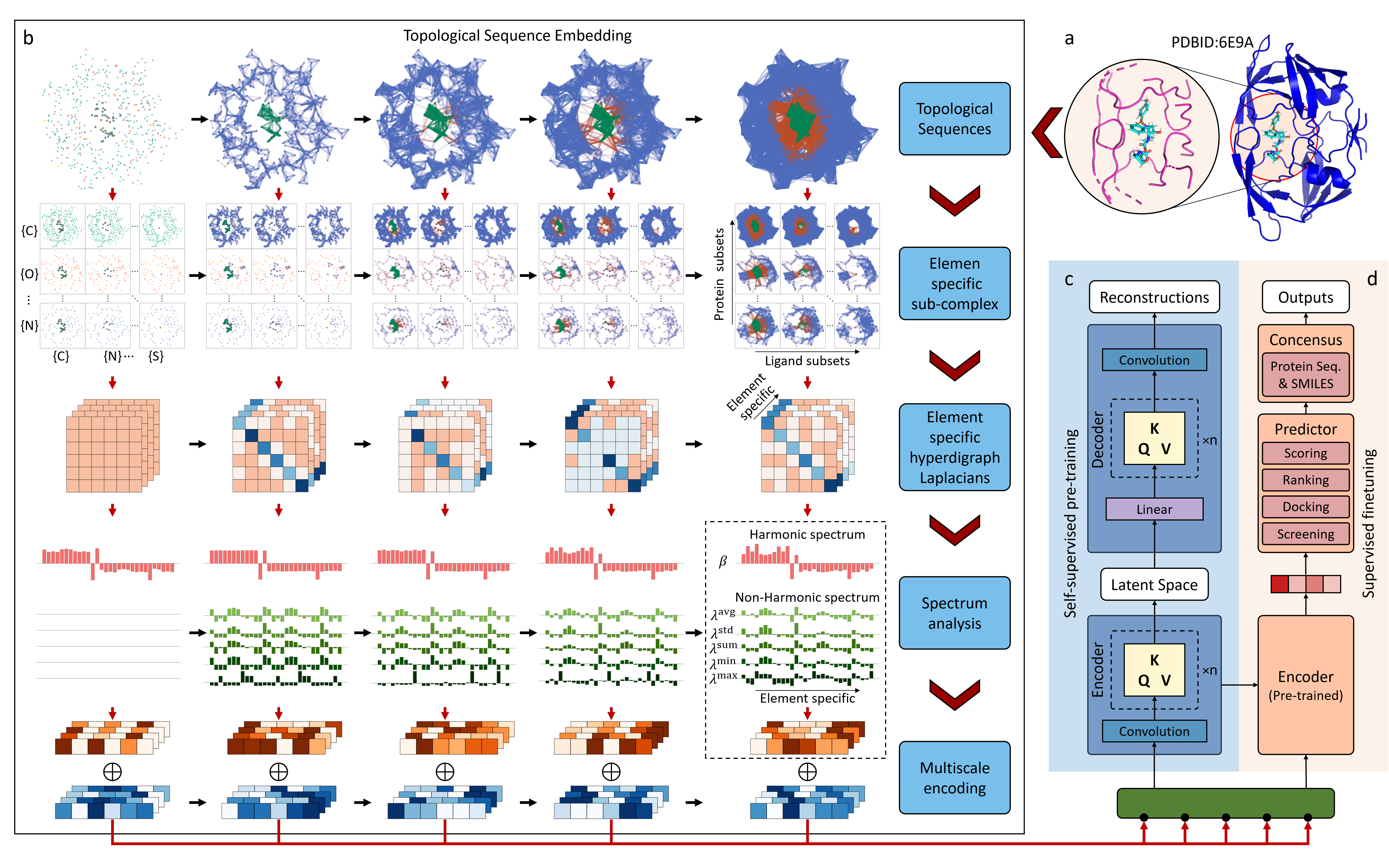
- TopoFormer
Topological Transformer (TopoFormer) is built by integrating NLP and a multiscale topology techniques, the persistent topological hyperdigraph Laplacian (PTHL), which systematically converts intricate 3D protein-ligand complexes at various spatial scales into a NLP-admissible sequence of topological invariants and homotopic shapes. Element-specific PTHLs are further developed to embed crucial physical, chemical, and biological interactions into topological sequences. TopoFormer surges ahead of conventional algorithms and recent deep learning variants and gives rise to exemplary scoring accuracy and superior performance in ranking, docking, and screening tasks in a number of benchmark datasets. The proposed topological sequences can be extracted from all kinds of structural data in data science to facilitate various NLP models, heralding a new era in AI-driven discovery. Keywords: Drug design, Topological sequences, Topological Transformer, Multiscale Topology, Hyperdigraph Laplacian.
Further explain the details in the paper, providing context and additional information about the architecture and its components.
- transformers 4.24.0
- numpy 1.21.5
- scipy 1.7.3
- pytorch 1.13.1
- pytorch-cuda 11.7
- scikit-learn 1.0.2
- python 3.9.12
git clone https://github.com/WeilabMSU/TopoFormer.git
| Datasets | Training Set | Test Set | |
|---|---|---|---|
| Pre-training | Combind PDBbind | 19513 RowData, TopoFeature_small, TopoFeature_large | |
| Finetuning | CASF-2007 | 1105 Label | 195 Label |
| CASF-2013 | 2764 Label | 195 Label | |
| CASF-2016 | 3772 Label | 285 Label | |
| PDB v2016 | 3767 Label | 290 Label | |
| PDB v2020 | 18904 Label (exclude core sets) |
195 Label (CASF-2007 core set) |
|
| 195 (CASF-2013 core set) |
|||
| 285 (CASF-2016 core set) |
|||
| 285 (v2016 core set) |
- RowData: the protein-ligand complex structures. From PDBbind
- TopoFeature: the topological embedded features for the protein-ligand complex. All features are saved in a dict, which
keyis the protein ID, andvalueis the topological embedded features for corresponding complex. The downloaded file is .zip file, which contains two file (1)TopoFeature_large.npy: topological embedded features with a filtration parameter ranging from 0 to 10 and incremented in steps of 0.1 \AA; (2)TopoFeature_small.npy: topological embedded features with a filtration parameter ranging from 2 to 12 and incremented in steps of 0.2 \AA; - Label: the .csv file, which contains the protein ID and corresponding binding affinity in the logKa unit.
| Task | Datasets | Description |
|---|---|---|
| Screening | LIT-PCBA | 3D poses for all 15 targets. Download (13GB) |
| PDBbind-v2013 | 3D poses. Download from https://weilab.math.msu.edu/AGL-Score | |
| Docking | CASF-2007,2013 | 3D poses. Download from https://weilab.math.msu.edu/AGL-Score |
# get the usage
python ./code_pkg/main_potein_ligand_topo_embedding.py -h
# examples
python ./code_pkg/main_potein_ligand_topo_embedding.py --output_feature_folder "../examples/output_topo_seq_feature_result" --protein_file "../examples/protein_ligand_complex/1a1e/1a1e_pocket.pdb" --ligand_file "../examples/protein_ligand_complex/1a1e/1a1e_ligand.mol2" --dis_start 0 --dis_cutoff 5 --consider_field 20 --dis_step 0.1bs=32 # batch size
lr=0.00008 # learning rate
ms=10000 # max training steps
fintuning_python_script=./code_pkg/topt_regression_finetuning.py
model_output_dir=./outmodel_finetune_for_regression
mkdir $model_output_dir
pretrained_model_dir=./pretrained_model
scaler_path=./code_pkg/pretrain_data_standard_minmax_6channel_large.sav
validation_data_path=./CASF_2016_valid_feat.npy
train_data_path=./CASF_2016_train_feat.npy
validation_label_path=./CASF2016_core_test_label.csv
train_label_path=./CASF2016_refine_train_label.csv
# finetune for regression on one GPU
CUDA_VISIBLE_DEVICES=1 python $fintuning_python_script --hidden_dropout_prob 0.1 --attention_probs_dropout_prob 0.1 --num_train_epochs 100 --max_steps $ms --per_device_train_batch_size $bs --base_learning_rate $lr --output_dir $model_output_dir --model_name_or_path $pretrained_model_dir --scaler_path $scaler_path --validation_data $validation_data_path --train_data $train_data_path --validation_label $validation_label_path --train_label $train_label_path --pooler_type cls_token --random_seed 1234 --seed 1234;# script for no validation data and validation label
# docking and screening
bs=32 # batch size
lr=0.0001 # learning rate
ms=5000 # max training steps
fintuning_python_script=./code_pkg/topt_regression_finetuning_docking.py
model_output_dir=./outmodel_finetune_for_docking
mkdir $model_output_dir
pretrained_model_dir=./pretrained_model
scaler_path=./code_pkg/pretrain_data_standard_minmax_6channel_filtration50-12.sav
train_data_path=./train_feat.npy
train_label_path=./train_label.csv
train_valdation_split=0.1 # 1/10 of training data will be used for validation
# finetune for regression on one GPU
CUDA_VISIBLE_DEVICES=1 python $fintuning_python_script --hidden_dropout_prob 0.1 --attention_probs_dropout_prob 0.1 --num_train_epochs 100 --max_steps $ms --per_device_train_batch_size $bs --base_learning_rate $lr --output_dir $model_output_dir --model_name_or_path $pretrained_model_dir --scaler_path /$scaler_path --train_data $train_data_path --train_label $train_label_path --validation_data None --validation_label None --train_val_split 0.1 --pooler_type cls_token --random_seed 1234 --seed 1234 --specify_loss_fct 'huber';# replace with the proper pathes
model_path=./pretrained_model
feature_path=./topo_feature.npy # it contains the topo_feature_array, rather than the dict
scaler_path=./code_pkg/pretrain_data_standard_minmax_6channel_filtration50-12.sav
save_feature_path=./latent_feature.npy
latent_python_script=./code_pkg/final_generate_latent_features.py
python $latent_python_script --model_path $model_path --scaler_path $scaler_path --feature_path $feature_path --save_feature_path $save_feature_path --latent_type encoder_pretrain- Scoring
| Finetuned for scoring | Training Set | Test Set | PCC | RMSE (kcal/mol) |
|---|---|---|---|---|
| CASF-2007 result | 1105 | 195 | 0.837 | 1.807 |
| CASF-2007 small result | 1105 | 195 | 0.839 | 1.807 |
| CASF-2013 result | 2764 | 195 | 0.816 | 1.859 |
| CASF-2016 result | 3772 | 285 | 0.864 | 1.568 |
| PDB v2016 result | 3767 | 290 | 0.866 | 1.561 |
| PDB v2020 result | 18904 (exclude core sets) |
195 CASF-2007 core set |
0.853 | 1.295 |
| 195 CASF-2013 core set |
0.832 | 1.301 | ||
| 285 CASF-2016 core set |
0.881 | 1.095 |
Note, there are 20 TopoFormers are trained for each dataset with distinct random seeds to address initialization-related errors. And 20 gradient boosting regressor tree (GBRT) models are subsequently trained one these sequence-based features, which predictions can be found in the results folder. Then, 10 models were randomly selected from TopoFormer and GBDT models, respectively, the consensus predictions of these models was used as the final prediction result. The performance shown in the table is the average result of this process performed 400 times.
- Docking
| Finetuned for docking | Success rate |
|---|---|
| CASF-2007 result | 93.3% |
| CASF-2013 result | 91.3% |
- Screening
| Finetuned for screening | Success rate on 1% | Success rate on 5% | Success rate on 10% | EF on 1% | EF on 5% | EF on 10% |
|---|---|---|---|---|---|---|
| CASF-2013 | 68% | 81.5% | 87.8% | 29.6 | 9.7 | 5.6 |
Note, the EF here means the enhancement factor. Each target protein has a finetuned model. result contains all predictions.
This project is licensed under the MIT License - see the LICENSE file for details.
If you use this code or the pre-trained models in your work, please cite our work.
- Chen, Dong, Jian Liu, and Guo-Wei Wei. "Multiscale topology-enabled structure-to-sequence transformer for protein–ligand interaction predictions." Nature Machine Intelligence (2024): 1-12. Read
# BibTex
@article{chen2024multiscale,
title={Multiscale topology-enabled structure-to-sequence transformer for protein--ligand interaction predictions},
author={Chen, Dong and Liu, Jian and Wei, Guo-Wei},
journal={Nature Machine Intelligence},
pages={1--12},
year={2024},
publisher={Nature Publishing Group UK London}
}This project has benefited from the use of the Transformers library. Portions of the code in this project have been modified from the original code found in the Transformers repository.
TopoFormer was developed by Dong Chen and is maintained by WeiLab at MSU Math