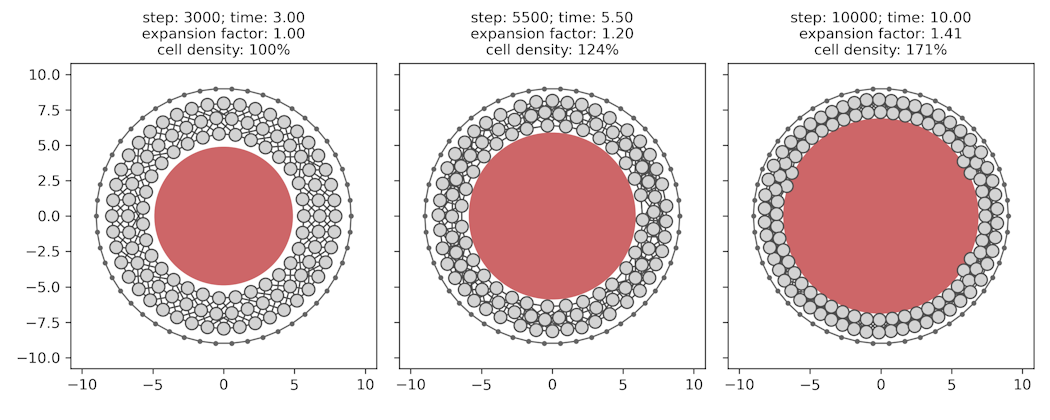
This repo hosts the code for the simple illustrative particle-based 2D model of cavity expansion and its effect on cell density for the paper "Competence for neural crest induction is controlled by hydrostatic pressure through YAP" by Alasaadi & colleagues. All code was written by Jonas Hartmann.
At each time step (simple_cell_sim.simulation.timestep), the pairwise distances between all cells are computed and converted into forces affecting each cell based on an anharmonic potential function. Cells thus strongly repulse each other at very short distances (resistance to compression), weakly attract each other at medium distances (adhesion), and do not affect each other at long distances. The positions of cells are then updated based on the summed contributions of all pairwise forces (and a small amount of Gaussian noise), scaled by the size of the time step (delta_t). The model is initialized as a circular band of uniformly spaced cells, with the outer-most ring of cells being fixed in place to represent the stiff vitelline membrane around the embryo. The cavity is modeled as a single large cell at the center that only imposes repulsive forces and whose resting size is increased over time to reflect growth of the cavity.
simple_cell_sim/simulation.py: functions for the simulation engine; the main one istimestep, which:- Retrieves pairwise distances between cells
- Computes the resulting forces for all provided force terms
- Adds a small amount of Gaussian noise
- Sums up the force contributions
- Updates the cell positions
simple_cell_sim/potential_funcs.py: functions relating distances between cells to potential energies- Only used for illustrative purposes in the current implementation
simple_cell_sim/force_funcs.py: functions relating distances between cells to forces- The ones used in the cavity model are
f_anharmonic(for cells) andf_Hooke(for the cavity)
- The ones used in the cavity model are
Notebooks/MODEL - Cavity Expansion.ipynb: jupyter notebook containing the cavity expansion model- Sets up the initial conditions and parameters
- Runs the simulation
- Produces figures & movie.
- To reproduce what is shown in the paper, simply install python and the required dependencies (see below) and run this notebook.
- Python
3.9.7and jupyter notebook (I used and recommend using the Anaconda distribution) - Scientific python packages
numpy==1.21.5matplotlib==3.5.3
- The study's lead author is Roberto Mayor (University College London). Please contact him for any queries about the paper.
- For code-specific question, you may contact Jonas Hartmann (University College London) or open an issue on GitHub.
- Note that I cannot promise support for any use cases other than direct reproduction of the study's results.