Knowledge Based Representation Learning for Nucleus Instance Classification from Histopathological Images
This repository contains PyTorch code for the paper: Knowledge Based Representation Learning for Nucleus Instance Classification from Histopathological Images
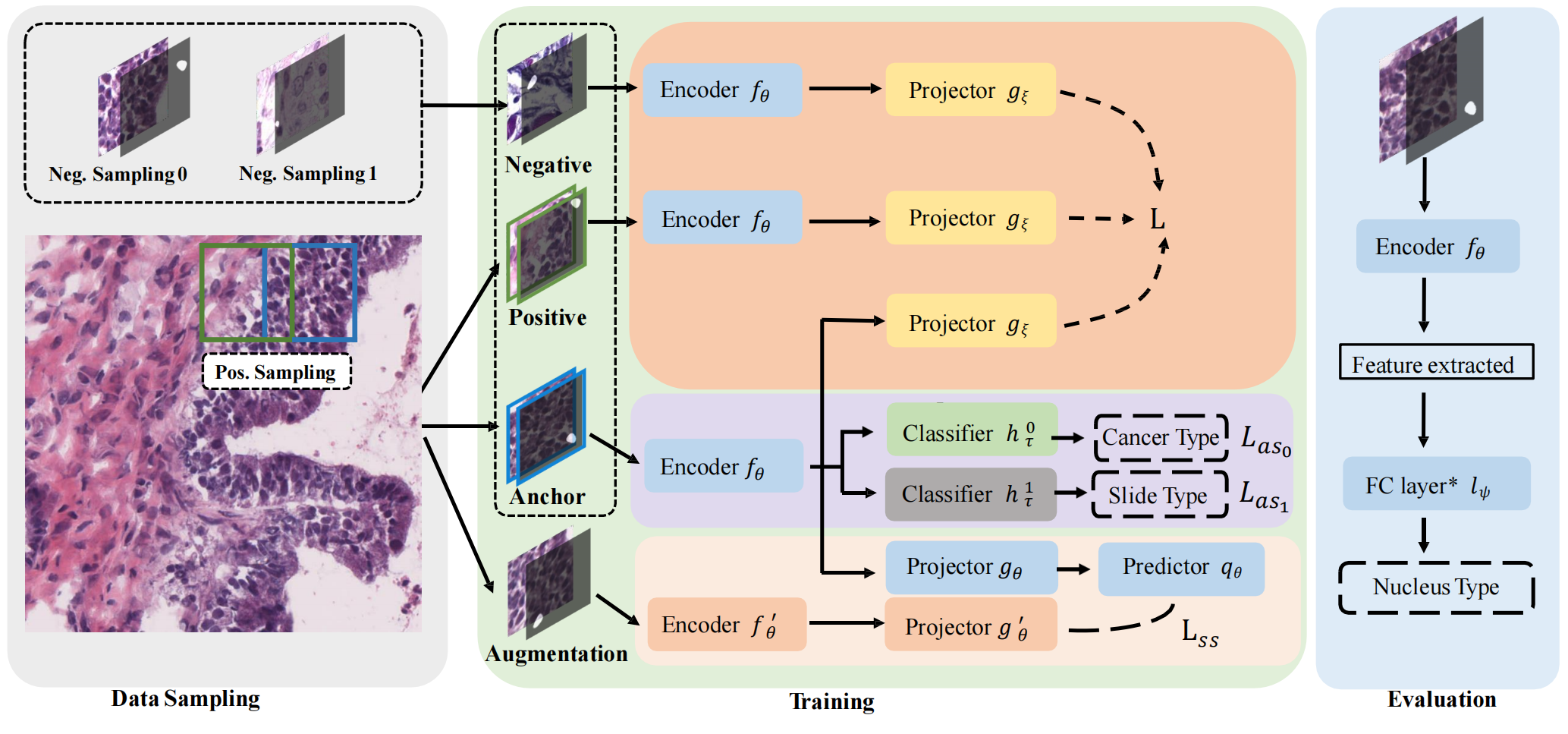
In this paper, we proposed a new framework for nucleus representation learning. We propose a new four-channel structured input and design Structured Triplet based on this input. We also add two auxiliary branches: the slide attribute learning branch and the conventional self-supervision branch, to further improve the representation encoder.
Links to the checkpoints can be found in the inference description below.
conda env create -n structured_triplet
conda activate structured_triplet
pip install torch torchvision
pip install tensorboard
pip install scikit-learn
pip install opencv
Below are the main directories in the repository:
dataset/: the data loader and augmentation pipelinetolls/: warm up training script and the main training scriptmodels/: model definitionutils/: defines the utility function
- Download our dataset and place it in a local accessible directory, e.g., /dataset/LarSPI
- Download the annotated dataset to evaluate on, e.g. panNuke or CoNSeP
Usage:
python tools/byol_warmup.py --root /dataset/LarSPI
python tools/train.py --root /dataset/LarSPI
Options:
--name The name of the experiment.
--byol_name The name of the warm up process
--lr Setting the learning rate.
--batch_size Setting the batch size.
--max_epoch Setting the epochs to train.
--alpha Setting the hyperparameter alpha
--beta Setting the hyperparameter beta
--resnet Setting the backbone of the framework
As part of our work, we provide the trained model at the link below:
python tools/train.py --root_eval /dataset/panNuke
Results of different classification methods on histopathological patches of 40 in PanNuke. (a) Input patch (b) SimCLR. (c) Moco (d)Moco v2 (e) BYOL (f) RCCNet (g) ViT (h) BiT (i) Structured-Triplet (j) Ground truth.
Our code on the byol branch is modified from byol-pytorch.
If any part of this code is used, please give appropriate citation to our paper.
@ARTICLE{9869632,
author={Zhang, Wenhua and Zhang, Jun and Yang, Sen and Wang, Xiyue and Yang, Wei and Huang, Junzhou and Wang, Wenping and Han, Xiao},
journal={IEEE Transactions on Medical Imaging},
title={Knowledge-Based Representation Learning for Nucleus Instance Classification from Histopathological Images},
year={2022},
volume={},
number={},
pages={1-1},
doi={10.1109/TMI.2022.3201981}}
The dataset provided here is for research purposes only. Commercial use is not allowed. The data is held under the following license: Attribution-NonCommercial-ShareAlike 4.0 International