Installation and usage instructions can be found in the wiki.
orchid
A management, annotation, and machine learning system for analyzing cancer mutations.
NOTE: This code is still an early release and is being actively developed. Please report any issues using the Issues tab and they will be fixed as soon as possible.
Introduction
Please refer to the following publication for a detailed description of this software:
Bioinformatics, btx709, https://doi.org/10.1093/bioinformatics/btx709
or, for a quick and dirty explanation:
What is orchid?
The purpose of orchid is to facilitate machine learning on tumor genetic data to gain biological or clinical insight. For example, you might be interested in sub-typing aggressive vs. non aggressive prostate cancer based on tumor mutational profiles derived from tumor sequence data, or maybe in trying to figure out which tumor tissue a cell-free DNA molecule is derived.
What is a 'tumor mutational profile'?
A tumor mutational profile is the annotated set of mutations within a tumor. A typical tumor might contain thousands of mutations, but most are assumed to be irrelevant to disease because they arise due to an important hallmark of cancer-- an unstable genome. These are called passenger mutations. However, some mutations (one to hundreds) may play important roles in carcinogenesis and/or be useful in identifying tumor characteristics, like aggressiveness. These are called driver mutations. Many cancer researchers focus only on driver mutations because of thier outsized role in cancer, but orchid takes the approach of analyzing all mutations in aggregate with machine learning algorithms to try to tease apart more subtle patterns. This approach makes sense since even mutations that have been deemed irrelevant have been associated with particular tumor types and may encode important information about (or even regulate processes involved in) the underlying biology of a tumor (e.g., trinucleotide signatures).
What is meant by an 'annotated set of mutations'?
An annotation is simply a numeric or ordinal value that can be associated with a particular mutation. For example, 'mutation A' may change the amino acid sequence of a protein, so we can annotate it as a 'non-synonymous single nucleotide polymorphism' or 'nsSNP'. On the other hand, 'mutation B' may not change the amino acid sequence, so we annotate it as a 'synonymous SNP'. Biologically speaking, a non-synonymous SNP is more likely to change the effect of a protein than a synonymous one. In machine learning parlance, an annotation is called a feature. If we gather many mutations across a tumor (or tumors) and annotate each mutation with many features, we end up with a set of annotated mutations, or tumor mutational profile.
At this time, many regulatory and coding features of the human genome have been extensively cataloged, resulting in a wealth of data to mine. If we gather enough biological data, we can increase our understanding of each individual mutation and its possible role in cancer, or at least begin to see if patterns emerge from the data. A list of features used in our publication and available in our public database can be found here (Note: This page is now only available from web.archive.org; actual file downloads possibly made on request): http://wittelab.ucsf.edu/orchid.
Here's an example. If we arrange a set of mutations from a tumor in rows and corresponding feature values in columns, a mutational profile can be created and visualized:
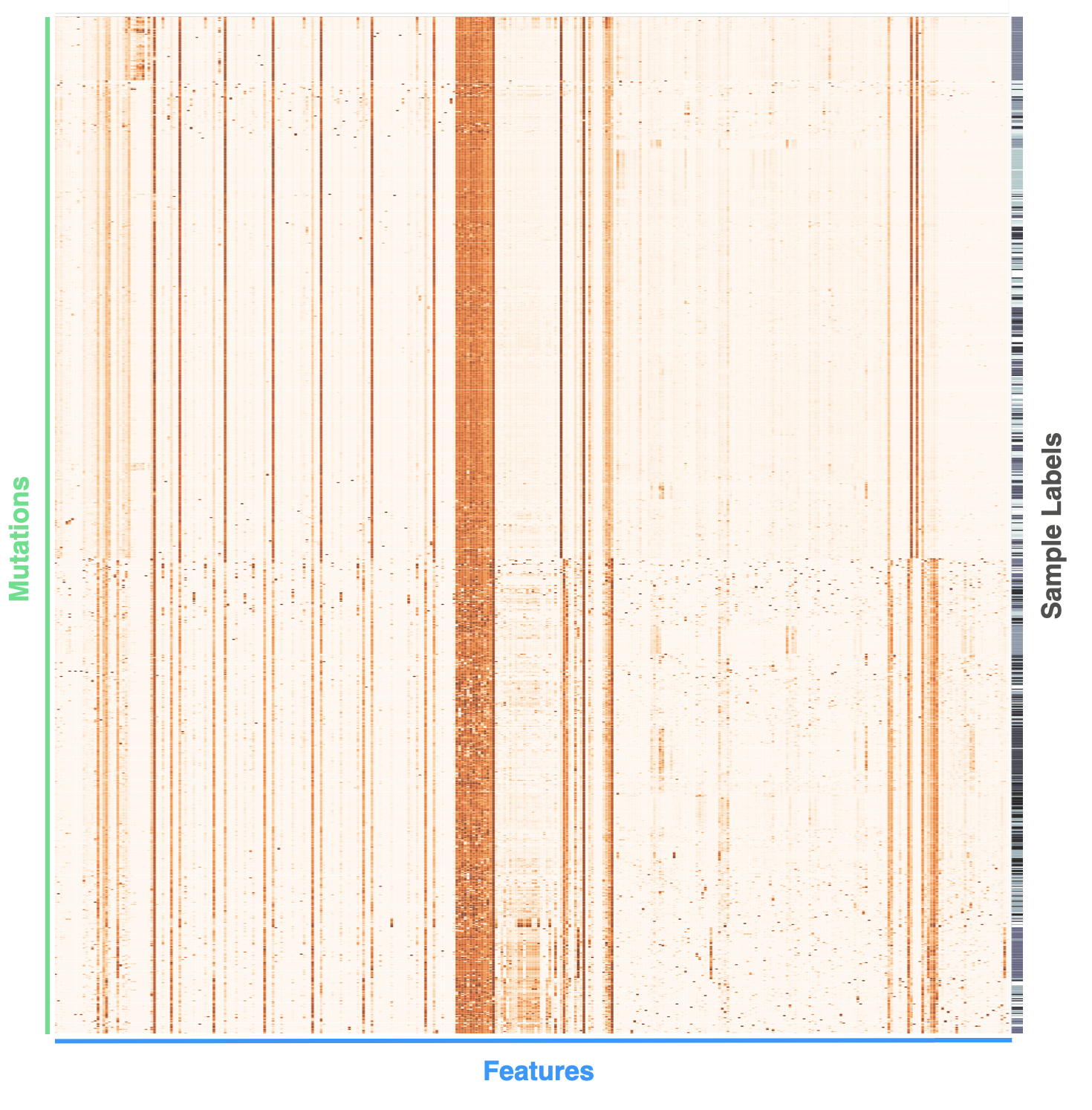
Here large feature values (or more 'severe' categories) are shown as more orange, while smaller (less 'severe') feature values are whiter. There is also a final column of sample labels, which is ultimately what we're interested in learning. In other words, this column's values are used to train supervised machine learning algorithms for the purpose of future sample classification.
Getting Started
- Download this code and install prerequisites
- Obtain tumor and annotation data
- Build the database
- Perform machine learning
Please refer to the wiki to begin!
NOTICE: This software requires the use of other code and/or data that must be obtained with respect to its license or copyright. Generally speaking, this implies orchid's use is restricted to non-commercial activities. Orchid itself is licensed under the MIT license requiring only preservation of copyright and license notices. Please see the LICENSE file for more details.
