Paper: https://arxiv.org/pdf/2302.03018.pdf
Please clone our environment using the following command:
conda env create -f environment.yml
conda activate DDM2
For fair evaluations, we used the data provided in the DIPY library. One can easily access their provided data (e.g. Sherbrooke and Stanford HARDI) by using their official loading script:
hardi_fname, hardi_bval_fname, hardi_bvec_fname = get_fnames('stanford_hardi')
data, affine = load_nifti(hardi_fname)Different experiments are controlled by configuration files, which are in config/.
We have provided default training configurations for reproducing our experiments. Users are required to change the path vairables to their own directory/data before running any experiments.
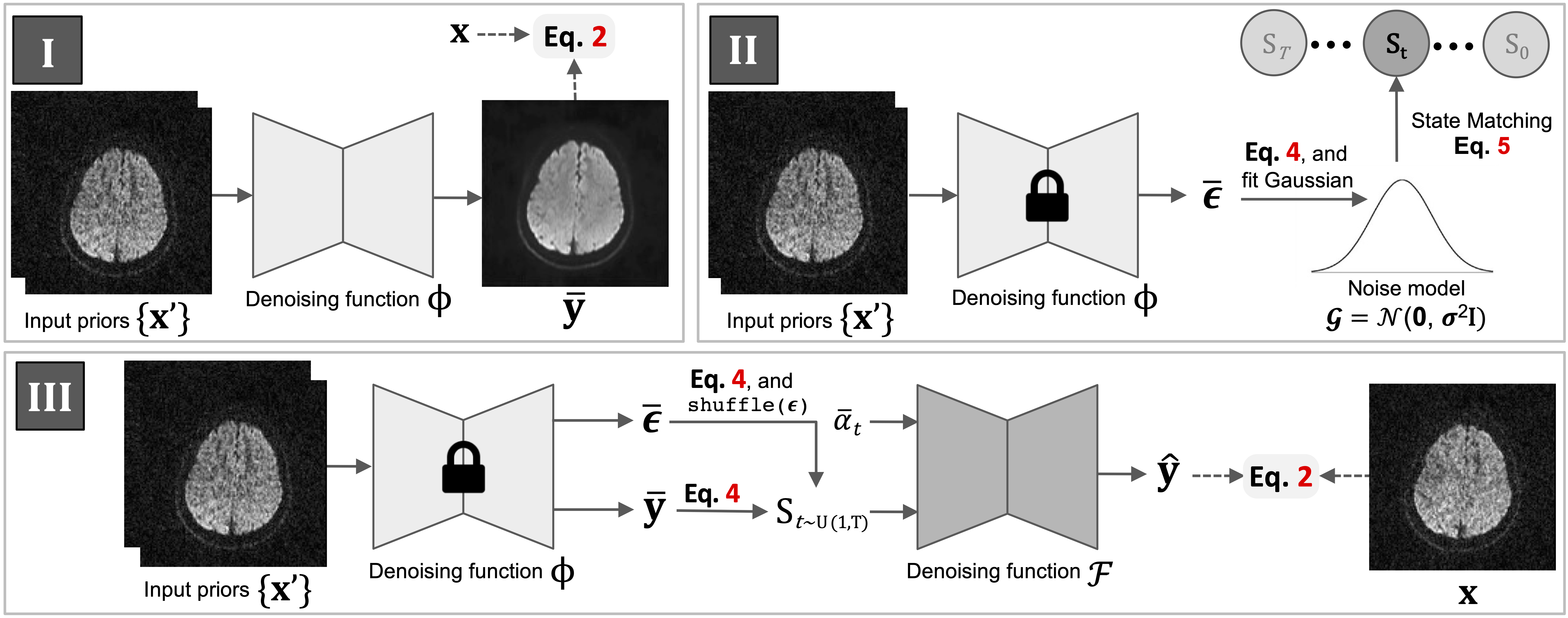
The training of DDM2 contains three sequential stages. For each stage, a corresponding config file need to be passed as a coommand line flag.
-
To train our Stage I:
python3 train_noise_model.py -p train -c config/hardi.json
or alternatively, modifyrun_stage1.shand run:
./run_stage1.sh -
After Stage I training completed, the path to the checkpoint of the noise model need to be specific at 'resume_state' of the 'noise_model' section in corresponding config file. Additionally, a file path (.txt) needs to be specified at 'initial_stage_file' in the 'noise_model' section. This file will be recorded with the matched states in Stage II.
-
To process our Stage II:
python3 match_state.py -p train -c config/hardi.json
or alternatively, modifyrun_stage2.shand run:
./run_stage2.sh -
After Stage II finished, the state file (recorded in the previous step) needs to be specified at 'initial_stage_file' for both 'train' and 'val' in the 'datasets' section.
-
To train our Stage III:
python3 train_diff_model.py -p train -c config/hardi.json
or alternatively, modifyrun_stage3.shand run:
./run_stage3.sh -
Validation results along with checkpoints will be saved in the
/experimentsfolder.
One can use the previously trained Stage III model to denoise a MRI dataset through:
python denoise.py -c config/hardi.json
or alternatively, modify denoise.sh and run:
./denoise.sh
The --save flag can be used to save the denoised reusults into a single '.nii.gz' file:
python denoise.py -c config/hardi.json --save
If you find this repo useful in your work or research, please cite:
@inproceedings{xiangddm,
title={DDM $\^{} 2$: Self-Supervised Diffusion MRI Denoising with Generative Diffusion Models},
author={Xiang, Tiange and Yurt, Mahmut and Syed, Ali B and Setsompop, Kawin and Chaudhari, Akshay},
booktitle={International Conference on Learning Representations}
}