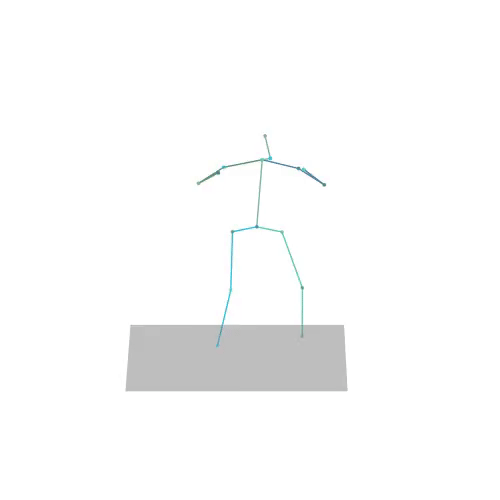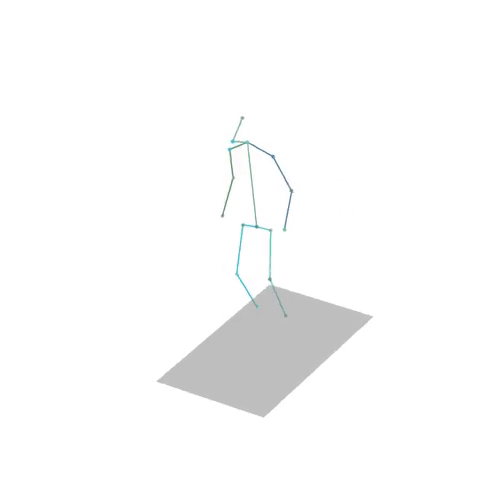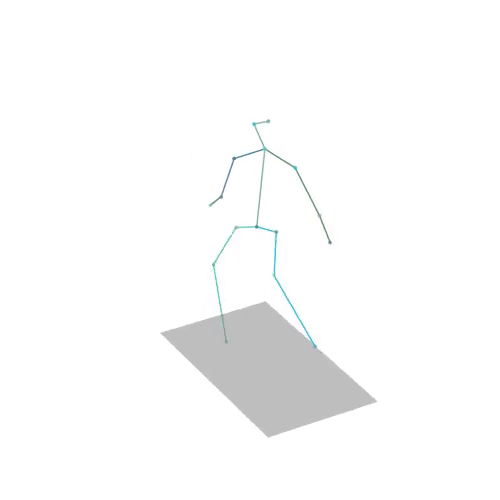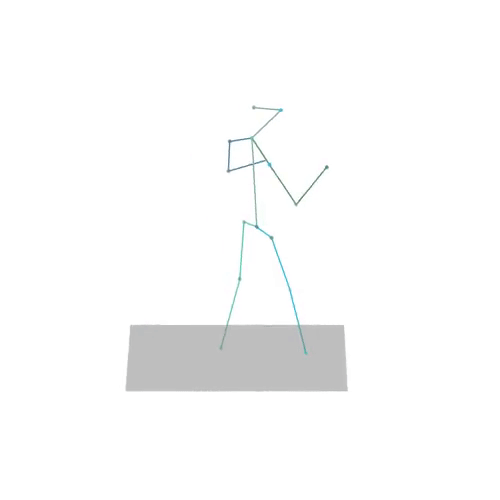
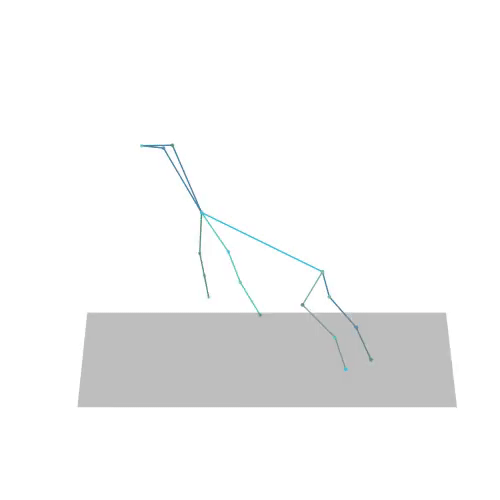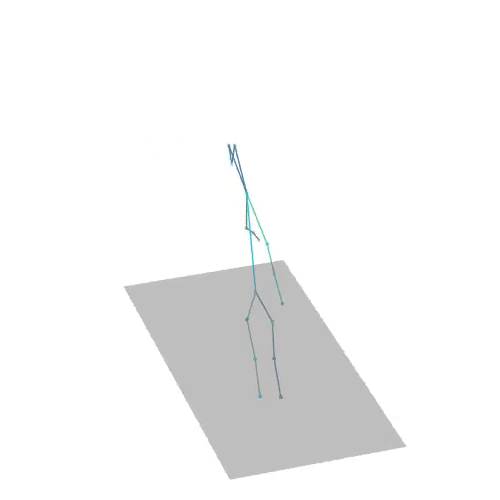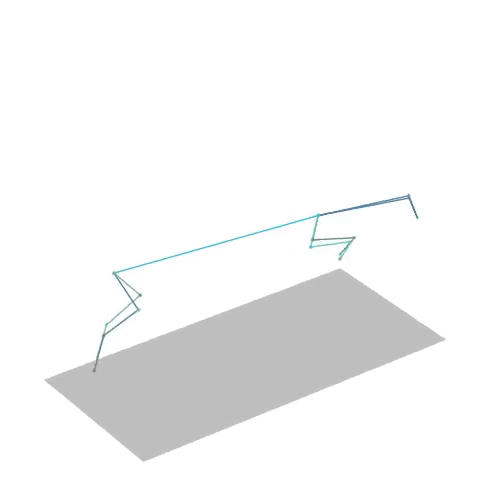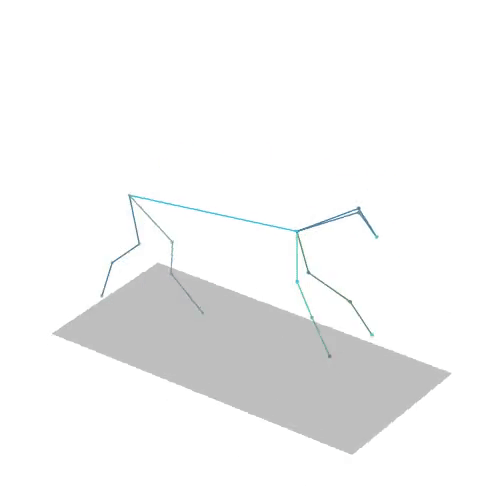
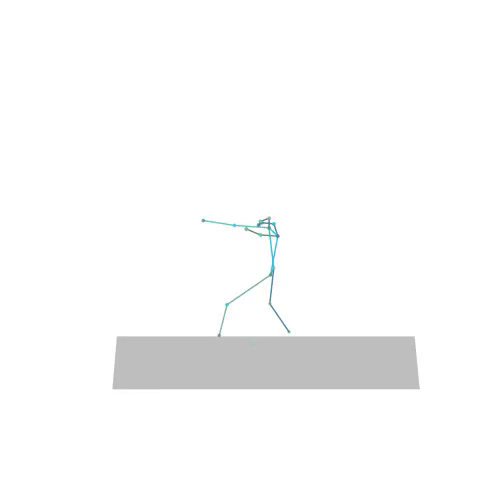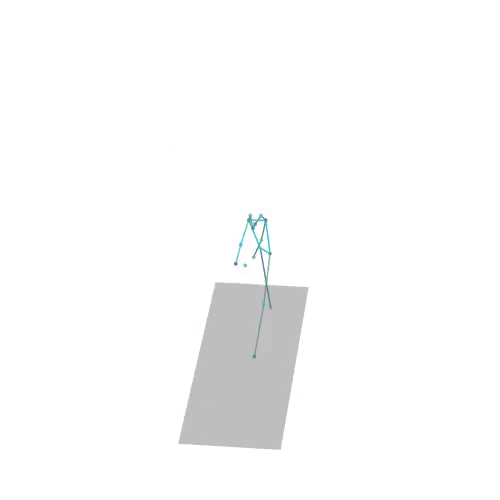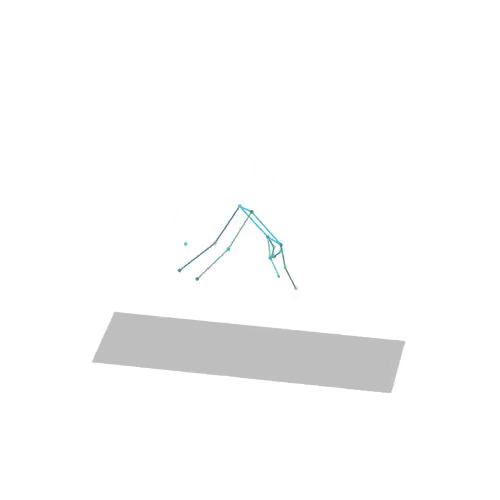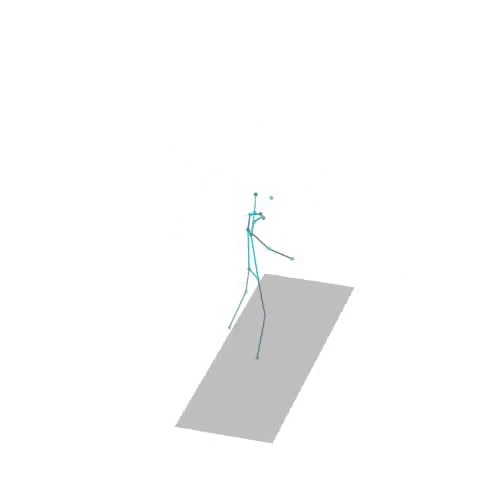
The official PyTorch implementation of the paper "MAS: Multi-view Ancestral Sampling for 3D motion generation using 2D diffusion".
📢 5/Nov/23 - First release
This code was tested on Ubuntu 18.04.5 LTS and requires:
- Python 3.7
- conda3 or miniconda3
- CUDA capable GPU (one is enough)
Setup conda env:
conda env create -f environment.yml
conda activate masIf you would like to to train or evaluate a model, you can download the relevant dataset. This is not required for using the pretrained models.
Download the zip file <dataset_name>.zip, then extract it into dataset/<dataset_name>/ inside the project directory. Alternatively, you can extract it to some other path and edit datapath in data_loaders/<dataset_name>/config.py to your custom path.
Note: NBA dataset is quite large in size so make sure you have enough free space on your machine.
3D pose estimations of the NBA dataset
You can download the results of 3D pose estimations by the supervised MotionBert and unsupervised Elepose on the 2D NBA dataset. You can later use them for evaluation, visualization, or for your own needs.
Download the zip <method>_predictions.zip file and unzip it into dataset/nba/<method>_predictions/. You can use an alternative path, and change the hard-coded paths in eval/evaluate.py (defined in the beginning of the file).
-
Using Elepose: elepose_predictions.zip
We supply the pretrained diffusion models for each dataset.
Download the model(s) you wish to use, then unzip them. We suggest to place them in save/<dataset>/.
We also supply the VAE model used for evaluation on the NBA dataset.
Download the model, then unzip it. We suggest to place it in save/evaluator/<dataset>.
Use the diffusion model to generate 2D motions, or use MAS to generate 3D motions.
General generation arguments
--model_path <path/to/checkpoint.pth>- the checkpoint file from which we load the model. The model's properties are loaded from theargs.jsonfile that is found in the same directory as the checkpoint file.--num_samples <number of samples>- number of generated samples (defaults to 10).--device <index>- CUDA device index (defaults to 0).--seed <seed>- seed for all random processes (defaults to 0).--motion_length <duration>- motion duration in seconds (defaults to 6.0).--use_data- use motion lengths and conditions sampled from the data (recommended, but requires downloading the data).--show_input_motions- plot the sampled 2D motions to a video file (only when specifying--use_data).--output_dir <directory path>- the path of the output directory. If not specified, will create a directory named<method_name>_seed_<seed>in the same directory as the<model_path>.--overwrite- if there already exists a directory with the same path as<output_dir>, overwrite the directory with the new results.
Run python -m sample.generate -h for full details.
python -m sample.generate --model_path save/nba/nba_diffusion_model/checkpoint_500000.pthNote: Make sure that the args.json file is in the same directory as the <model_path>.
python -m sample.mas --model_path save/nba/nba_diffusion_model/checkpoint_500000.pthMAS arguments
- All of the general generation arguments are available here.
--num_views <number of views>- number of views used for MAS (defaults to 7).
Run python -m sample.mas -h for full details.
Dynamic view-point sampling
Use MAS with new view angles sampled on each iteration (Appendix C in the paper).
python -m sample.ablations --model_path save/nba/nba_diffusion_model/checkpoint_500000.pth --ablation_name random_anglesAlternative methods
Generate a 3D motion with alternative methods.
Specify --ablation_name to control the type of method to apply. Can be one of [mas, dreamfusion, dreamfusion_annealed, no_3d_noise, prominent_angle, prominent_angle_no_3d_noise, random_angles].
python -m sample.ablations --model_path save/nba/nba_diffusion_model/checkpoint_500000.pth --ablation_name dreamfusionRunning one of the 2D or 3D motion generation scripts will get you a new directory <output_dir>, which includes:
results.npyfile with all conditions and xyz positions of the generated motions.result_<index>.mp4- a stick figure animation for each generated motion. 3D motions are plotted with a rotating camera, and repeat twice (this is why there is a sharp transition in the middle):
You can stop here, or generate an explicit mesh using the following script.
Generate a SMPL mesh
For human motions, it is possible to create a SMPL mesh for each frame:
Download dependencies:
bash prepare/download_smpl_files.shIf the script fails, create body_models/ directory, then download the zip file smpl.zip and extract it into the directory you created.
Then run the script:
python -m visualize.render_mesh --input_path /path/to/mp4/video/result_<index>.mp4 --skeleton_type <type>This script outputs:
<file_name>_smpl_params.npy- SMPL parameters (thetas, root translations, vertices and faces)<file_name>_obj/- A directory with a mesh per frame in.objformat.
Notes:
- Important - Make sure that the results.npy file is in the same directory as the video file.
- The
.objcan be integrated into Blender/Maya/3DS-MAX and rendered using them. - This script is running SMPLify and needs GPU as well (can be specified with the
--deviceflag). - You have two ways to animate the sequence:
- Use the SMPL add-on and the theta parameters saved to
<file_name>_smpl_params.npy(we always use beta=0 and the gender-neutral model). - A more straightforward way is using the mesh data itself. All meshes have the same topology (SMPL), so you just need to keyframe vertex locations.
Since the OBJs are not preserving vertex order, we also save this data to the
<file_name>_smpl_params.npyfile for your convenience.
- Use the SMPL add-on and the theta parameters saved to
Mesh generation arguments
--input_path </path/to/mp4/video/result_<index>.mp4to specify which sample to convert to a mesh.--skeleton_type <type>to specify the skeleton type of the motion. Can be one of [nba,motionbert,elepose,gymnastics].
python -m train.train_mdm --save_dir save/nba/my_nba_diffusion_model --dataset nbaTraining with evaluation
Add evaluation during training using the --eval_during_training flag.
python -m train.train_mdm --save_dir save/nba/my_nba_diffusion_model --dataset nba --eval_during_training --evaluator_path save/evaluator/nba/no_aug/checkpoint_1000000.pth --subjects "model mas" --num_views 5 --num_eval_iterations 2Training arguments
See Evaluate section for the evaluation arguments.
--save_dir <path>- the directory to save results to (required).--device <index>- CUDA device index (defaults to 0).--seed <seed>- seed for all random processes (defaults to 0).--lr <learning rate>- learning rate of the gradient decent algorithm (defaults to 10^(-5)).--resume_checkpoint <path/to/checkpoint.pth- resume training from an existing checkpoint.--train_batch_size- batch size during training (defaults to 32). If the algorithm exits due to lack of memory, you can reduce the batch size.--save_interval <interval>- number of optimization steps applied between each checkpoint saving. Make sure not to decrease this one too much since the models can take up a lot of space. If you also run an evaluation it could also slow down the training process.--train_platform_type {ClearmlPlatform, TensorboardPlatform}- track results with either ClearML or Tensorboard. Make sure to have follow the appropriate setup for your platform before running this.--overwrite_model- if the model<save_dir>already exists, overwrite it with the new results.
Data arguments
--dataset <dataset name>- which dataset to use (defaults to"nba"). Most other data arguments are automatically loaded depending on this argument, but can be overridden, or permanently changed indata_loaders/<dataset_name>/config.py.--data_augmentations <list of space-separatated data augmentations>- apply some data augmentations. The supported augmentations are [shift_aug,scale_aug,length_aug] (default loaded from dataset config). We recommend to uselength_aug. If you wish to later apply an evaluation on your model, we advise not to use the augmentations since otherwise it would yield worse results.--datapath <path>- the dataset path (default loaded from dataset config).--data_size <size>- select a random subset of the dataset in the specified size. This can critically reduce loading time if you only need a small subset. If not specified, the entire dataset will be loaded.--data_split <split>- which data split to use. Can be one of [train,test] (defaults to"train").
Note: If you wish to add your own dataset, you would have to implement a dataset class (recommended to inherit from BaseDataset), create a config.py file and add your dataset to the list in data_loaders/dataset_utils.py. See the existing datasets for reference.
Model arguments
--arch <architecture>- model architecture. Can be one of [trans_enc,trans_dec,gru] (defaults totrans_enc).--layers <number of layers>- number of attention layers (defaults to8).--latent_dim <dimension size>- latent dimension size of the transformer (defaults to512).--num_heads <number of heads>- number of attention heads in the attention layers (defaults to4).--dropout <dropout rate>- dropout rate during training (defaults to0.1).--activation <activation function>- activation function in the feed forward layer (defaults to"gelu").--ff_size <feed forward size>- size of feed forward internal dimension (defaults to1024).--emb_trans_dec- inject condition as a class token (fortrans_decarchitecture only).
Diffusion arguments
--model_mean_type <type>- what the model is trained to predict. Can be one of [x_start,epsilon] (defaults tox_start). MAS supports epsilon prediction but we did not extensively test it.--noise_schedule <schedule>- the noise schedule of the diffusion process. Can be one of [linear,cosine,cosine_tau_<exponenet>] (defaults tocosine_tau_2). Thecosine_tau_<number>simply means taking the cosine schedule and to the power of<exponent>.--diffusion_steps <number of steps>- number of diffusion steps (defaults to100).--sigma_small <True/False>- variance type of the backwards posterior (defaults toTrue, which means a small variance).
Running one of the training scripts will get you a new directory named <save_dir>/, which includes:
args.json- a file with the model's configuration.checkpoint_<step>.pth- checkpoint files that includes the model's and optimizers' state at the given training step.
Evaluate 3D motion by projecting it to random angles and comparing the resulting 2D distribution to the 2D test data distribution. For more details refer to the paper.
python -m eval.evaluate --evaluator_path save/evaluator/nba/nba_evaluator/checkpoint_1000000.pth --model_path save/nba/nba_diffusion_model/checkpoint_500000.pth --subjects "ElePose motionBert model mas train_data" --num_views 5If you did not download elepose and motionBert's predictions, you can remove them from the subject list.
Evaluation arguments
--subjects <list of space-separated method names>- which methods to apply the evaluation on. Each subject can be one of [motionBert,ElePose,train_data,model,mas,dreamfusion,no_3d_noise].--evaluator_path <path/to/checkpoint.pth>- evaluator checkpoint to use.--angle_mode <mode>- angle distribution during evaluation. Can be one of [uniform,side,hybrid] (defaults touniform).--eval_num_samples- number of samples used in each evaluation iteration. If you would like to compare to our reported results, do not change it (defaults to1024).--num_eval_iterations- number of repititions of the experiment (defaults to10).
Run python -m eval.evaluate -h for full details.
Running the evaluation script will get you a file named <model_path>/checkpoint_<step>_eval_<angle_mode>.txt:
- The results are organized in a table where the columns are the metric types and the rows are the evaluation subjects.
- Each cell includes the mean value of the metric for the tested subject across all experiment repititions, with confidence intervals of 95%, denoted by
±<interval_size/2>.
Evaluator training
python -m eval.train_evaluator --save_dir save/evaluator/nba/my_nba_evaluator --dataset nbaAll of the training and data arguments from also apply here.
Evaluator model arguments
--e_num_layers <number of layers>to control how many attention layers the model uses (defaults to6).--e_latent_dim <dimension size>to specify the latent dimension size of the transformer (defaults to256).--e_num_heads <number of heads>to specify the number of attention heads (defaults to4).--e_dropout <dropout rate>to set the dropout rate during training (defaults to0.1).--e_activation <activation function>to control which activation function is used (defaults to"gelu").--e_ff_size <feed forward size>to set the feed forward internal size (defaults to1024).
Our code heavily relies on the intense labor and dedication of others, and we would like to thank them for their contribution:
MDM, guided-diffusion, MotionCLIP, text-to-motion, actor, joints2smpl, MoDi.
This code is distributed under an MIT LICENSE.
Note that our code is based on other libraries, including CLIP, SMPL, SMPL-X and PyTorch3D.
If you find this code useful in your research, please cite:
@misc{kapon2023mas,
title={MAS: Multi-view Ancestral Sampling for 3D motion generation using 2D diffusion},
author={Roy Kapon and Guy Tevet and Daniel Cohen-Or and Amit H. Bermano},
year={2023},
eprint={2310.14729},
archivePrefix={arXiv},
primaryClass={cs.CV}
}