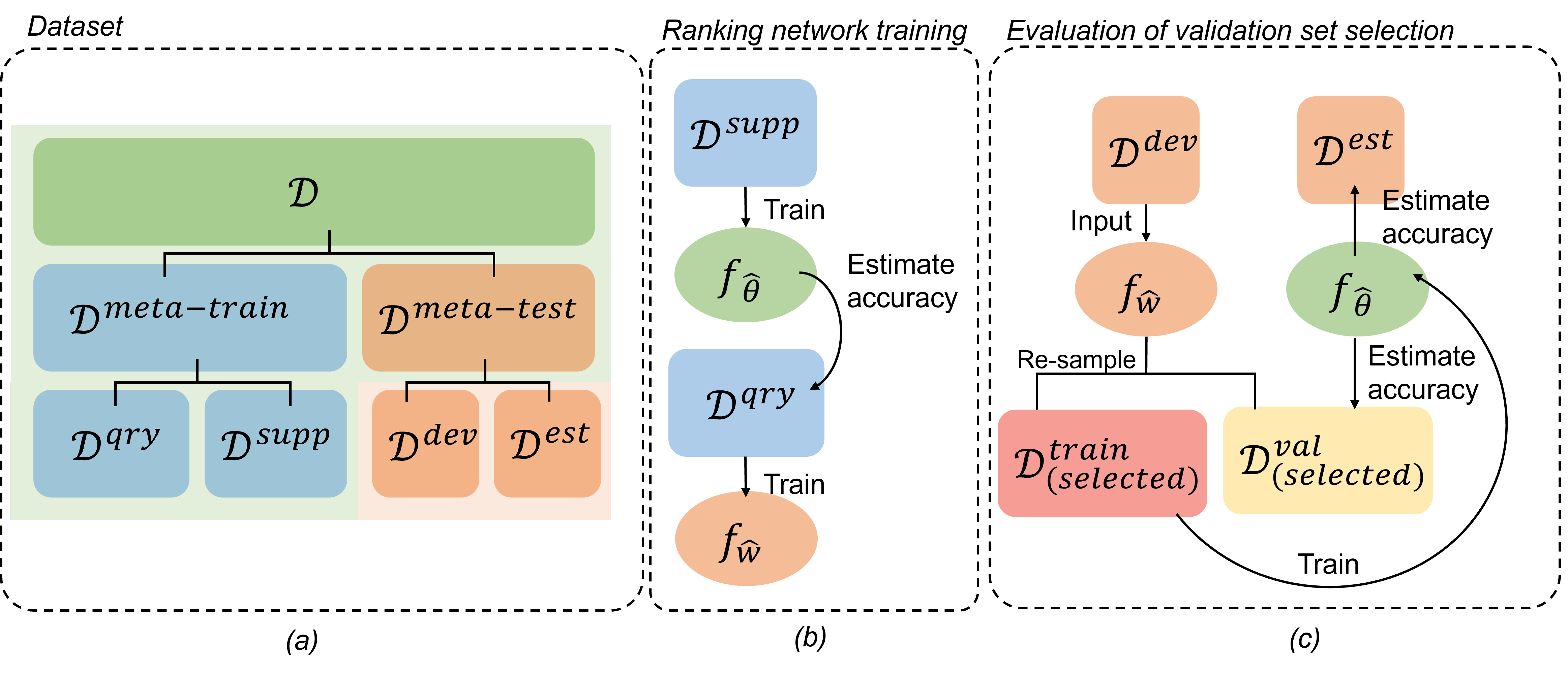
Selecting performance-representative validation sets for developing medical image segmentation neural networks
Code accompanying the paper
## Recommended Requirements ``` Python == 3.11.4, PyTorch == 2.0.1, monai == 1.2.0 ``` NOTE: Other versions of Python and Pytorch may still compatible with this repo but not guaranteed.Instructions for acquiring PTB and WT2 can be found here. Download and unzip to the root dir. All data examples is cropped and store into a h5 file for quicker load time. In the jupyer notebook file data_preparation/pre_process_prostate.ipynb, simply call the function "convert_h5(original data dir, destination data dir)" with two input strings as described. One example's image and label will be processed and store into one file with unique index.
Simply run and output a txt file with shuffled data dir name so that spilt of dataset remain consistent.
python data_preparation data_dir_shuffle.py
Train segmentation network from scratch, run
python train_seg_only.py cuda:0 4 your_seg_nick_name
Train ranking network from scratch, run
python train_sel_only.py cuda:0 6 your_seg_nick_name
Simply run
python post_hold_out_120
If you use any part of this code in your research, please cite our paper:
@article{XXX
}