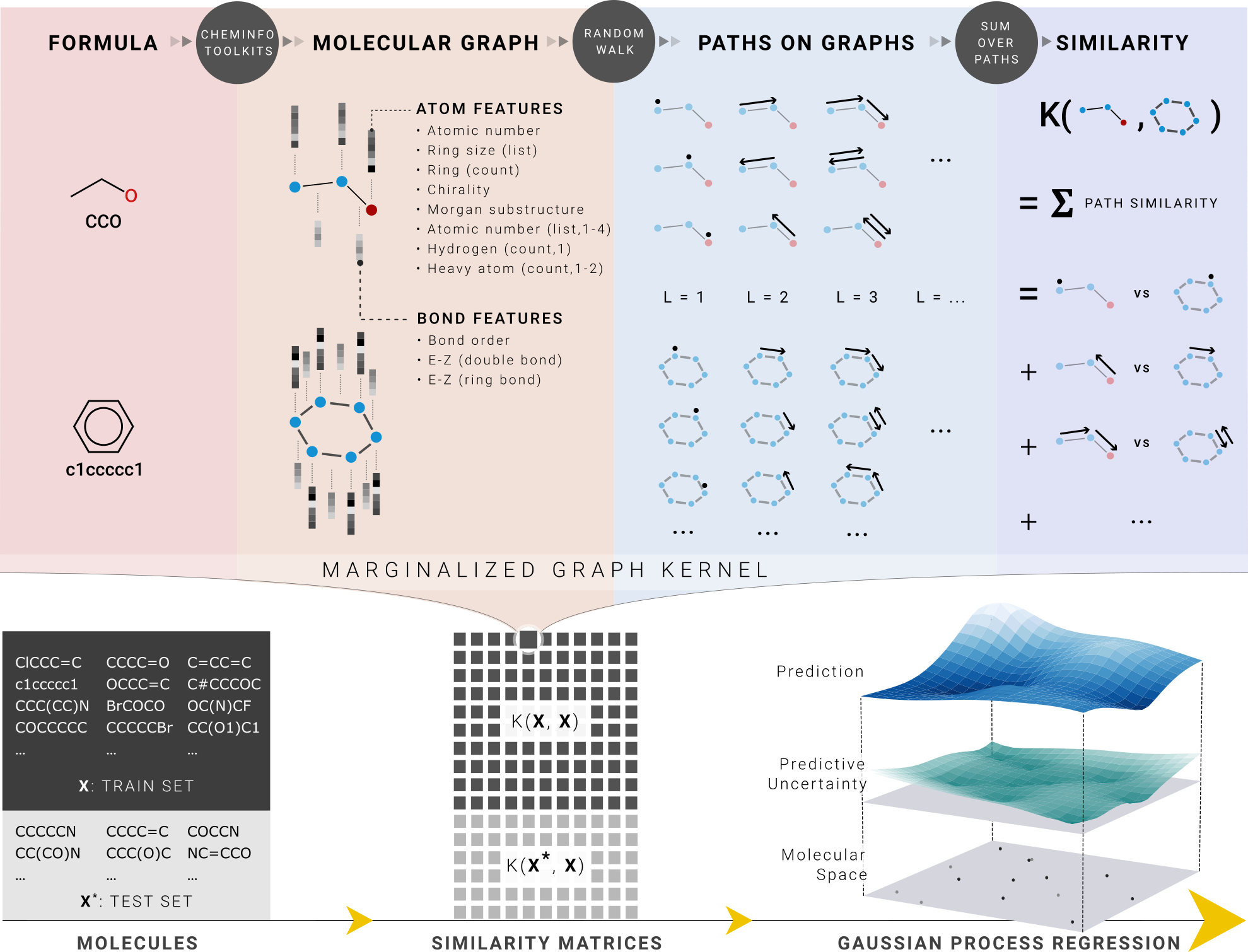
Predicting molecular properties using Marginalized Graph Kernel, GraphDot.
It supports regression (GPR) and classification (GPC, SVM) tasks on
- pure compounds.
- mixtures.
Besides molecular graph, additional vector could also be added as input, such as temperature, pressure, etc.
Predicting Single-Substance Phase Diagrams: A Kernel Approach on Graph Representations of Molecules.
A Comparative Study of Marginalized Graph Kernel and Message-Passing Neural Network.
GCC (7.*), NVIDIA Driver and CUDA toolkit(>=10.1). Python 3.10 is suggested.
pip install numpy==1.22.3 git+https://gitlab.com/Xiangyan93/graphdot.git@feature/xy git+https://github.com/bp-kelley/descriptastorus typed-argument-parser mgktools==1.1.0
For some combinations of GCC and CUDA, only old version of pycuda workspip install pycuda==2020.1
- The executable files are in directory run.
- The hyperparameter files in json format are placed in directory hyperparameters.