SCeQTL is an R package that uses zero-inflated negative binomial regression to do eQTL analysis on single-cell data. It can distinguish two type of gene-expression differences among different genotype groups. It’s more suitable to use SCeQTL to identify eQTLs from single-cell data. It can also be used for finding gene expression variations associated with other grouping factors like cell lineages. Following is the detail information and usage of this program. You can also found them in READ.ME. R Packages is a book based around this workflow.
Hu Yue y-hu10@qq.com
Xi Xi xix19@mails.tsinghua.edu.cn
To install the developmental version from GitHub (recommended):
if(!require(devtools)) install.packages("devtools")
devtools::install_github("XuegongLab/SCeQTL")To install from source package, you could download the R package from Mac and Linux version or Windows version and run the command in R:
install.packages(<source_code_file>, repos=NULL, type="source") qvalue
ggplot2
doParallel
pscl(<=1.5.2)
SCeQTL takes two inputs: gene and snp.
The input gene is a scRNA-seq raw read counts matrix. The rows of the matrix are genes and columns are cells.
The other input snp is a genotype matrix, where each element should be 0/1/2 indicating three types of genotypes. The order of columns should be the same with gene.
Users can load the test data in SCeQTL by
library(SCeQTL)
data(test)
The toy data gene in test is a scRNA-seq read counts matrix which has 151 genes (rows) and 1529 cells (columns).
dim(gene)
gene[1:6, 1:6]
The object snp in test is a matrix of genotype which has three levels and equal length to the column number of snp.
dim(snp)
snp[1:6, 1:6]
Here is an example to run SCeQTL with read counts matrix input:
# Load library and the test data for SCeQTL
library(SCeQTL)
data(test)
# check whether the non-zero part could fit negative binomial well
# This function may fail since It's possible that the random picked gene can't be fit to negative bionomial distribution, all zero value for example
checkdist(gene)
# normalize gene matrix using DEseq method
normalize(gene)
# Detecting the SCeQTL
result <- cal.pvalue(gene, snp)
# Picking one sample to visualize
check.sample(gene[1,], snp[1,])
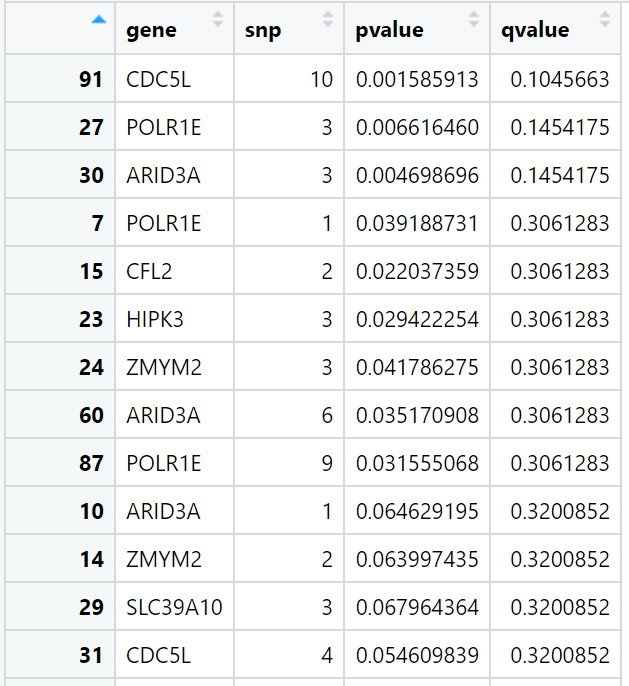
SCeQTL will output the p-value of each gene-snp pair. You could also get q-value by running cal.qvalue(gene, snp) instead of cal.pvalue(gene, snp).
SCeQTL::cal.qvalue(gene[1:10,], snp)
The result is like the picture below.
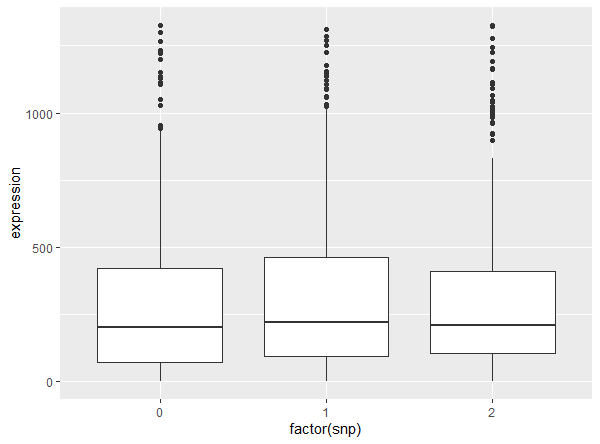
Then you may interested in the significant gene-snp pairs. You could stat and visualize it by check.sample. Here take the most significant pair for example.
SCeQTL::check.sample(gene["CDC5L",], snp[10, ])
Then you could get the statistic about the pair and a boxplot showing the relationship.
zero ratio of group 0 0.02004008
zero ratio of group 1 0.006944444
zero ratio of group 2 0
mean of group 0: 286.8818
mean of group 1: 302.4861
mean of group 2: 318.8277
non-zero part mean of group 0: 292.7485
non-zero part mean of group 1: 304.6014
non-zero part mean of group 2: 318.8277
non-zero part standard error of group 0: 282.2443
non-zero part standard error of group 1: 292.6688
non-zero part standard error of group 2: 288.2995
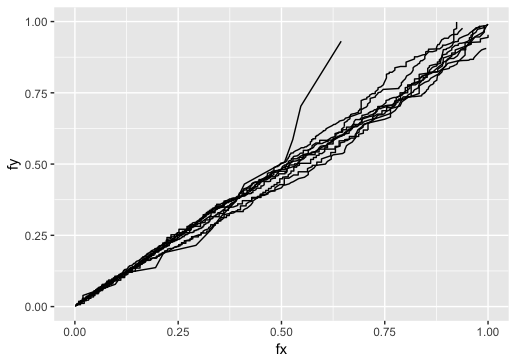
Before run cal.qvalue, you may want to check whether the distribution fit the bionomial negative assumption well by checkdist
checkdist(gene)
A Q-Q plot will be shown. It will randomly pick some genes so you may get different kind of result.
Use the following code in R to get access to the help documentation for DEsingle:
# Documentation for checking and preparing gene expression dataset
?checkdist
?normalize
# Documentation for finding single cell eQTL
?cal.pvalue
?cal.qvalue
# Documentation for checking single gene-snp pair
?check.sample
# Documentation for TestData
?test
?snp
?group
You are also welcome to contact the author by email for help.
Please note that the SCeQTL project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.