Algorithm for finding gene spatial pattern based on Gaussian process accelerated by SOM
conda install -c conda-forge somoclu
conda install pandas
conda install patsy
pip install somdeSlide-seq data we used can be downloaded from SpatialDB website: http://www.spatialomics.org/SpatialDB/download.php
df = pd.read_csv(dataname+'count.csv',sep=',',index_col=1)
corinfo = pd.read_csv(dataname+'idx.csv',sep=',',index_col=0)
corinfo["total_count"]=df.sum(0)
X=corinfo[['x','y']].values.astype(np.float32)After data loading, we can generate a SOM on the tissue spatial domain.
from somde import SomNode
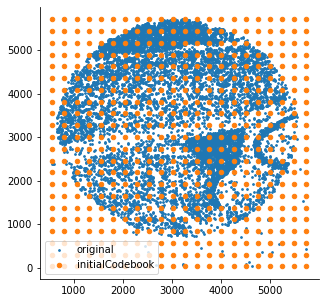
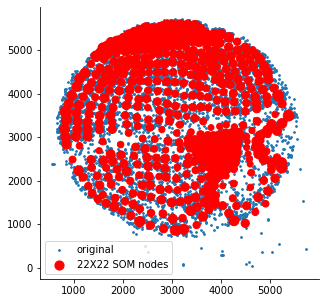
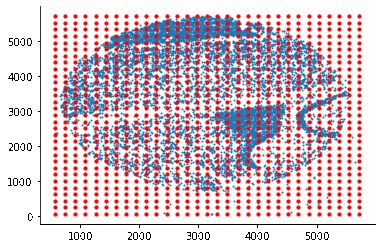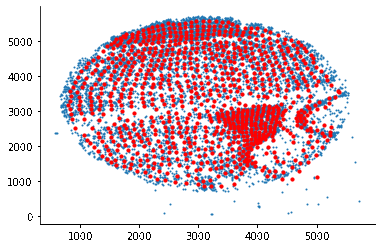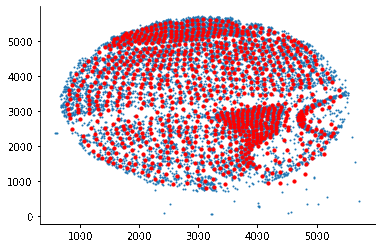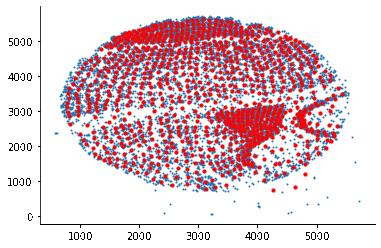
som = SomNode(X,20)This step will initialize a SOM and train it. You can use som.viewIniCodebook()andsom.view() to visualize the distribution of untrained and trained SOM nodes. Here is the result:
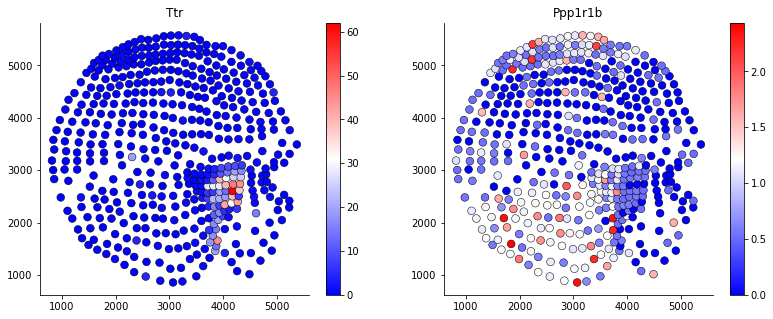
ndf,ninfo = som.mtx(df)mtxfunction will generate pesudo gene expression and spatial data site information at reduced resolution. You can visualize the gene expreesion on the condensed map by using plotgene.
Since we integrated the original count data, we need to normalize gene expression matrix in each SomNode object.
nres = som.norm()
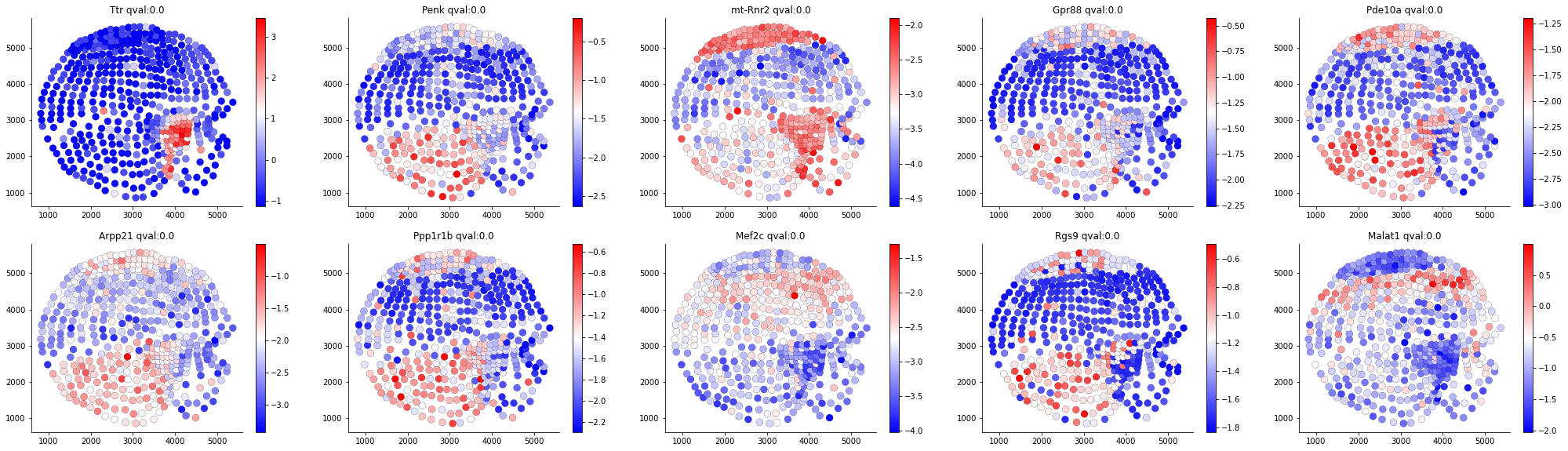
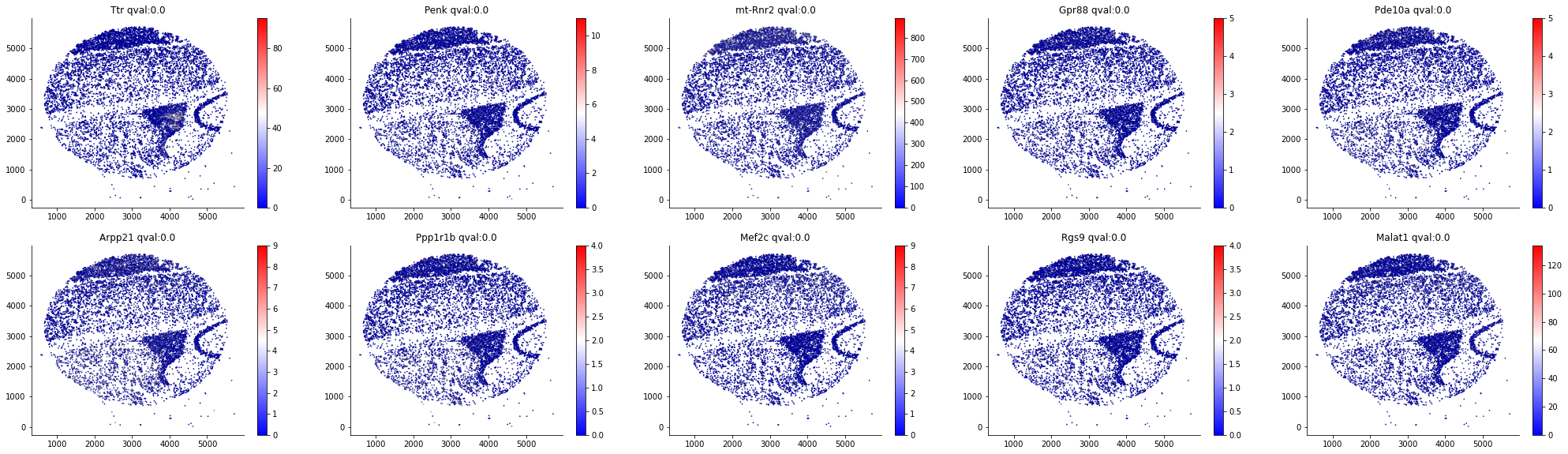
result, SVnum =som.run()The identification step is mainly based on the adjusted Gaussian Process, which was first proposed by SpatialDE. Here are the Top SVgenes given by SOMDE on both the original and SOM view:
More visualization results can be found at https://github.com/WhirlFirst/somde/blob/master/slide_seq0819_11_SOM.ipynb