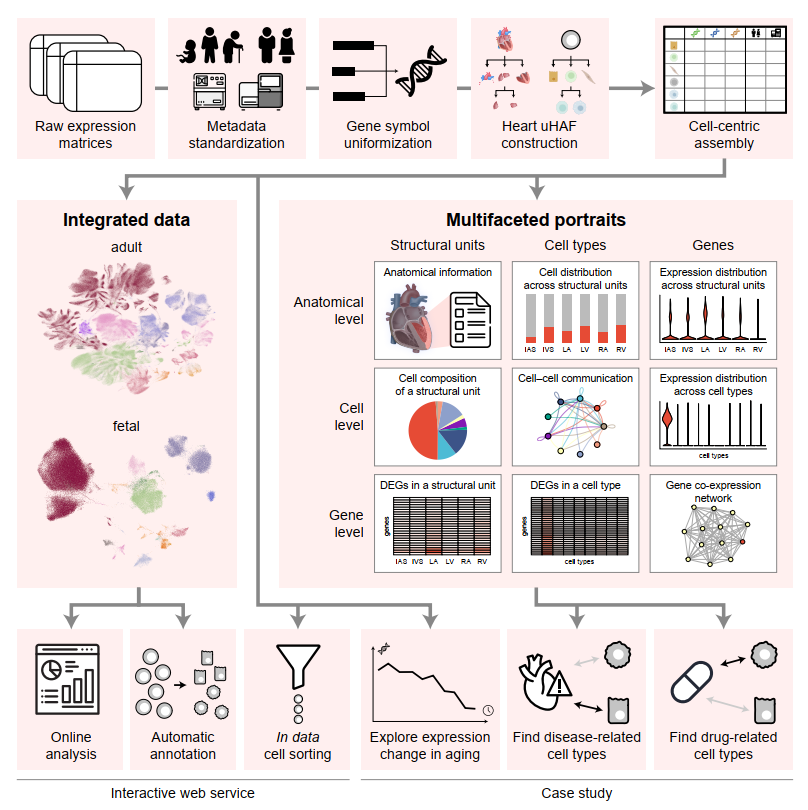
We collected scRNA-seq and snRNA-seq data of healthy human hearts from all available sources and built the first human ensemble heart cell atlas, uniHEART, using a unified information framework for cell-centric atlas assembly. uniHEART is available at https://heart.unifiedcellatlas.org/.
This repository contained the codes of:
- data processing
- integration benchmarking experiments
- data integration
- construction of multifacted portraits
- scripts of case studies
- online analysis tool
- ECAUGT2 tool
For the analysis of data and construction of portraits, we used following software: R=4.0.3, CellChat=1.1.0, Seurat=4.0.0, WGCNA=1.70.3, DEGseq=1.44.0, limma=3.46.0, GSVA=1.38.2, clusterProfiler=3.18.1, variancePartition=1.20.0. For data integration, we used following software: python=3.9.12 + torch=1.12.1(cuda 12.0), scanpy=1.9.1, trVAE=1.1.2, bbknn=1.3.9, scvi-tools=0.16.2, scgen=2.1.0, scanorama=1.7, scikit-learn=1.2.2, R=4.0.3, Seurat=4.1.1, SeuratWrappers=0.3.0, and harmony=0.1.0. For the development of ECAUGT2 and the Multinomial Dirichlet model for proportion estimation, we used following software: python=3.9.12 + torch=1.11.0(cuda 11.4), scipy=1.8.1, and scikit-learn=1.1.1.