This repository contains the code used to perform the analysis and generate the figures in this paper:
Benchmarking long-read RNA-sequencing analysis tools using in silico mixtures Xueyi Dong, Mei R. M. Du, Quentin Gouil, Luyi Tian, Jafar S. Jabbari, Rory Bowden, Pedro L. Baldoni, Yunshun Chen, Gordon K. Smyth, Shanika L. Amarasinghe, Charity W. Law, Matthew E. Ritchie Nature Methods 2023; doi: https://doi.org/10.1038/s41592-023-02026-3
Our RNA-seq data are available from Gene Expression Omnibus (GEO) under accession number GSE172421 (main benchmarking dataset) and GSE227000 (lab-based mixture of replicate 1).
Please cite our paper if you use our data and/or scripts in your studies.
All scripts are available at pilot (Illumina) and pilot_ONT/scripts (ONT)
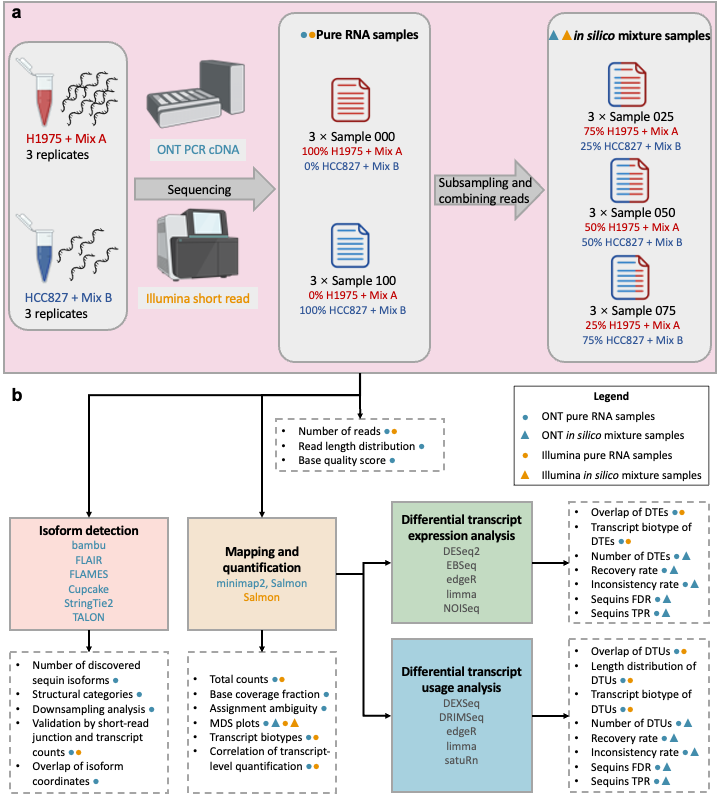
ONT: ONT/mix_prepare
Illumina: illumina/mix_prepare
Scripts are available at downsample
ONT: ONT/preprocess
Illumina: illumina/salmon_map.sh
ONT-specific: ONT/QC
General: longvsshort/overdisp.R, longvsshort/qc.R and longvsshort/sequinCPMvsAbundance.R
Scripts to run softwares: ONT/isoform_detection/methods
Analysis of results: ONT/isoform_detection/analysis
ONT: ONT/DE_mix.Rmd
Illumina: illumina/DE_mix.Rmd
Results comparison: longvsshort/DEmixres.R
ONT: ONT/DTU_mix.Rmd
Illumina: illumina/DTU_mix.Rmd
Results comparison: longvsshort/DTUmixres.R and longvsshort/DTUmixrestx.R