The last version is here: LINK
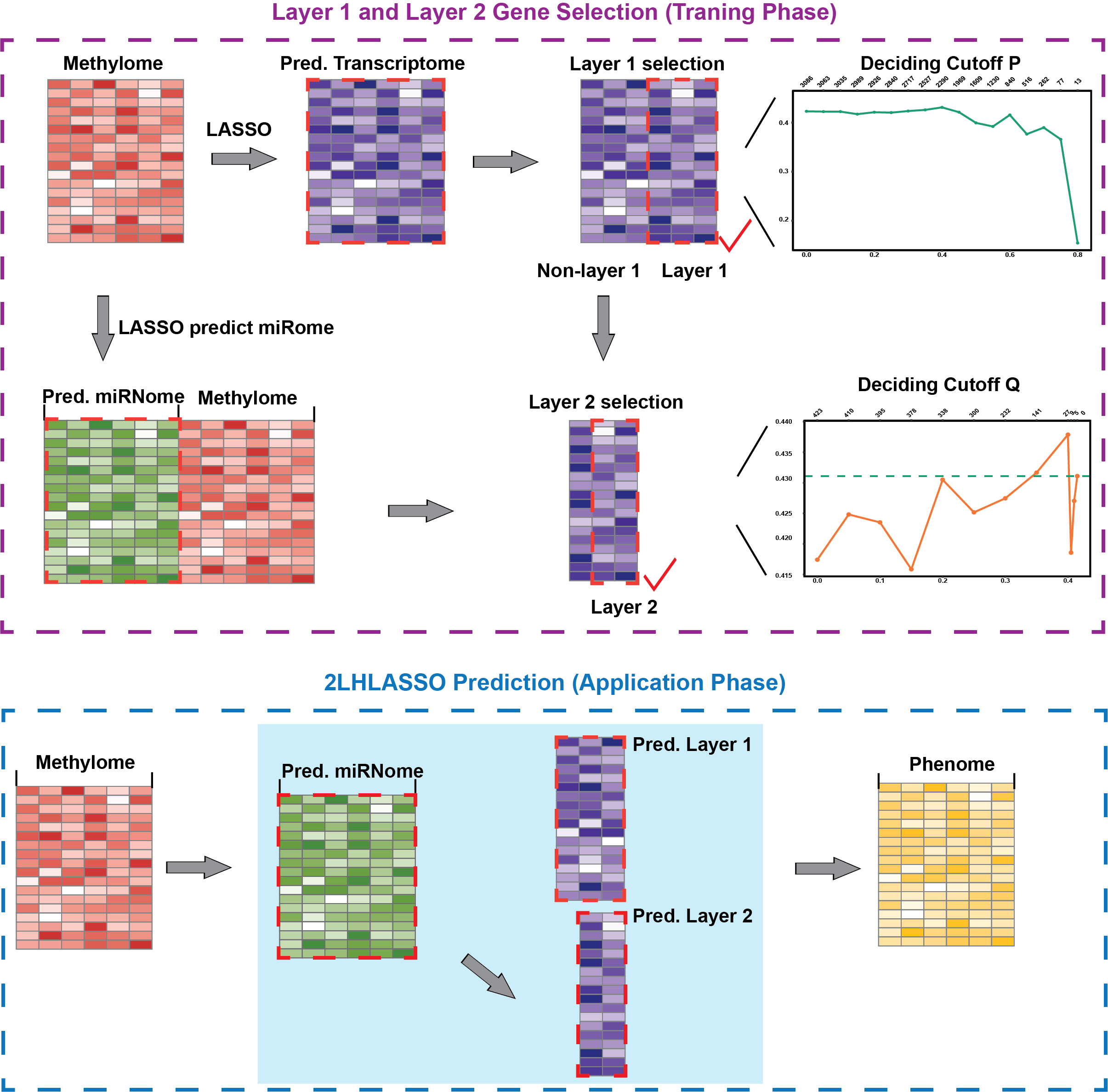
Two-layer hierarchical LASSO (2LHLASSO) instrumental variable methodology to integrate multi-omics predictor variables based on the flow of genetic information in the central dogma. Now we mainly apply the 2LHLASSO model on prostate cancer cohort to make the prediction of prostate cancer outcome. The Gleason score (GS), considered the most relevant clinical indicator for biochemical relapse-free survival (BRFS), was treated as the target outcome trait to be predicted, and the DNA methylation profiles were employed as the instrumental variables to estimate the expected transcription profiles. The analysis of the TCGA prostate cancer dataset showed that the new method of 2LHLASSO had higher predictability for outcome traits when compared to the linear model that simply combined multi-omics variables.
Simply clone this repository and the main example is in script: main.R
You will need the following R packages installed to run TLHLASSO:
- dplyr
- tidyr
- glmnet
- DT
In this model, GS was treated as the targeted quantitative trait to be predicted, and the DNA methylation profiles were employed as the instrumental variables.
-
In step 1, the methylomic data were used to estimate the expected (or predicted) expression profiles for the mRNAs in layer 1.
-
In step 2, we again used the methylomic data to estimate the expected expression levels for miRNAs, in accordance with the fact that methylation also regulates the abundance of miRNAs.
-
In step 3, for the mRNAs in layer 2 which initially had lower methylome-based predictabilities, we recalculated their expected expression values in step 3 with improved predictabilities by using both methylomic data and the predicted abundance of miRNAs. This was based on the knowledge that both methylation and miRNAs can modulate genomic transcription.
-
In step 4, we applied the LASSO model with cross-validation to rescreen mRNAs in both layers based on their expected values and used these fine-tuned mRNA profiles in the final model for the prediction of GS.
- R_square.R: The function to calculate predictability.
- cross_lasso.R: The function to conduct the cross validation LASSO regression and predict the response variable.
- load_data_and_divide_group.R: The script to load the example dataset and generate the group for cross validation.
The example of TLHLASSO model is in file main.R. Please run line by line to get the results.