---Yalong Liu---
Liu, Yalong, Jie Li, Miaomiao Wang, Zhicheng Jiao, Jian Yang, and Xianjun Li. "Trident Segmentation CNN: A Spatiotemporal Transformation CNN for Punctate White Matter Lesions Segmentation in Preterm Neonates." arXiv preprint arXiv:1910.09773 (2019). Arxiv Paper
Key words: Deep Learning; Punctate white matter lesions (PWMLs); Brain tumor segmentation.
This repository(~1.5MB) includes:
1.T1WI of 10 patients for test. (Full dataset is not allowed to be made public)
2.Full code for model training and inference.
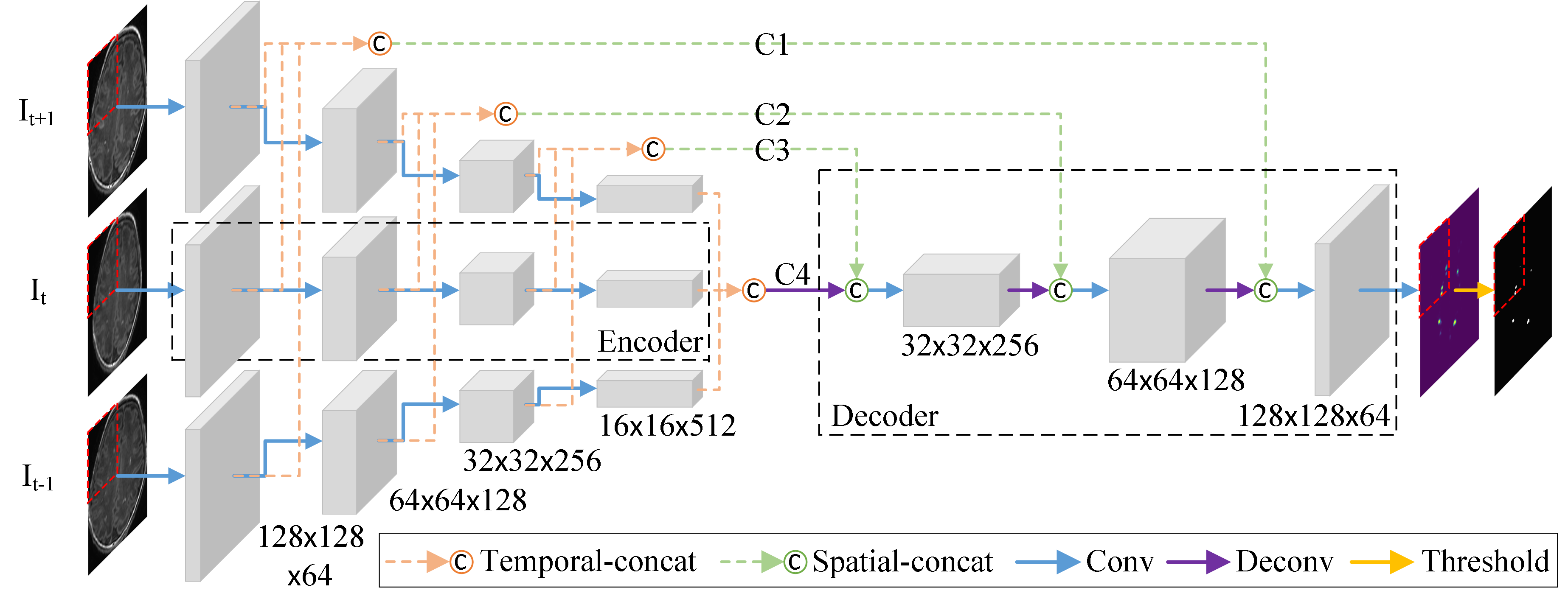
1.Trident Segmentation CNN (Convert spatial information into temporal information, which reduces the consumption of computing resources.)
Fig. 1. A Deep Learning method to segment punctate white matter lesions (PWMLs).
2.Self-balancing Focal Loss (Automatically balancing the loss of different classes in class imbalance situation and boost the model performance.)
Where SBFL0 and SBFL1 are the focal loss of background and foreground pixels without α . sum(SBFL0) and sum(SBFL1) are the sums of SBFL0 and SBFL1 . So sum(SBFL0)+sum(SBFL0) represents the total loss of the model. Intuitively, we can use the ratio of SBFL0 and sum(SBFL0)+sum(SBFL0) to balance the loss of SBFL0 and SBFL1 . However, this may result in a sharp oscillation of the model parameters. In order to segment PWML, we will focus on the segmentation of the lesion areas when balancing the loss of positive and negative samples. That is, β should always be greater than 0.5. It also should ensure that the model does not only focus on the segmentation of positive areas, so we limit the maximum value of β to 0.9 by a coefficient of 0.4. Finally, the SBFL is composed of SBFL1 weighted by β and SBFL0 weighted by 1-β.(For more information, please refer to my Arxiv paper)
Python 3.6.3
Tensorflow-gpu 1.15.2
CUDA 10.0
(Tested on windows 10)
- Download pretrained weights(~257MB) (BaiDuYun (Password: aivi), GoogleDrive). and put it in the './logs' folder.
- Choose a mode in the main.py('inference' or 'training') and run the code.
(Optional) You can modify parameters in './configs/pwml/config_TridentSegNet.py' according to the comments.
This repo borrows tons of code from
matterport/Mask_RCNN
1.Trident Segmentation CNN:
| N | Model | DSC | Sensitive | Specificity | Hausdorff Distance |
Notes |
|---|---|---|---|---|---|---|
| A | Mukherjee et al. [4] | 0.4288 | 0.4533 | 0.9961 | 59.6432 | A Triditional Method |
| B | Residual U-Net [7] | 0.5838 | 0.6013 | 0.9998 | 44.7323 | Residual U-Net (patch-based) |
| C | RS-RCNN [2] | 0.5986 | 0.6535 | 0.9998 | 36.6065 | Refined Segmentation R-CNN |
| D | TS-CNN | 0.6024 | 0.6838 | 0.9998 | 30.9768 | Proposed |
| E | TS-CNN (Data-aug) | 0.6088 | 0.6838 | 0.9998 | 30.8133 | + data augmentation |
| F | TS-CNN (CC=64) | 0.6186 | 0.6952 | 0.9998 | 28.3413 | Double channel counts to 64 |
| G | TS-CNN (SBFL) | 0.6355 | 0.7126 | 0.9998 | 24.5836 | Trained by Self-balancing Focal Loss |
2.Self-balancing Focal Loss:
Fig. 2. SBFL can automatically balance the class0 loss and class1 loss in the training process to get better performance.
Table 1. Results on MR images
| N | Original MR images |
Mukherjee et al. [4] | Residual U-Net [7] | RS-RCNN [2] | Our Method (TS-CNN) |
|---|
| 1 | 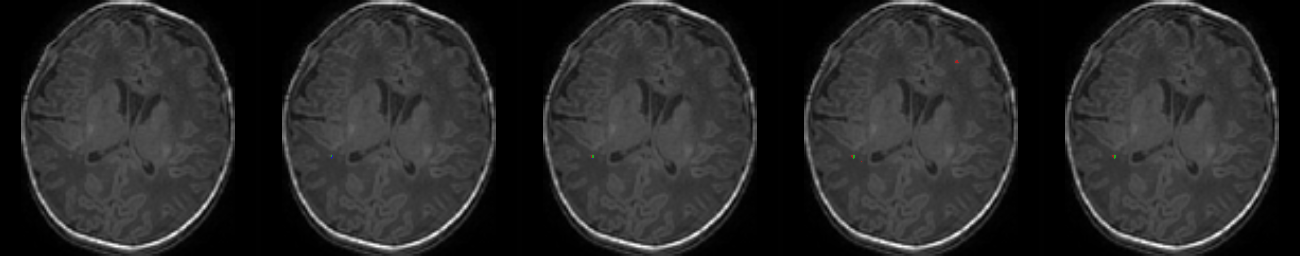 |
|---|---|
| 2 | 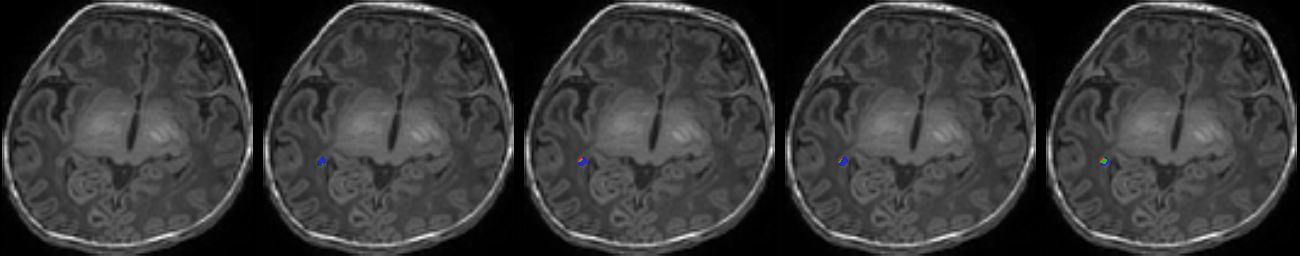 |
| 3 | 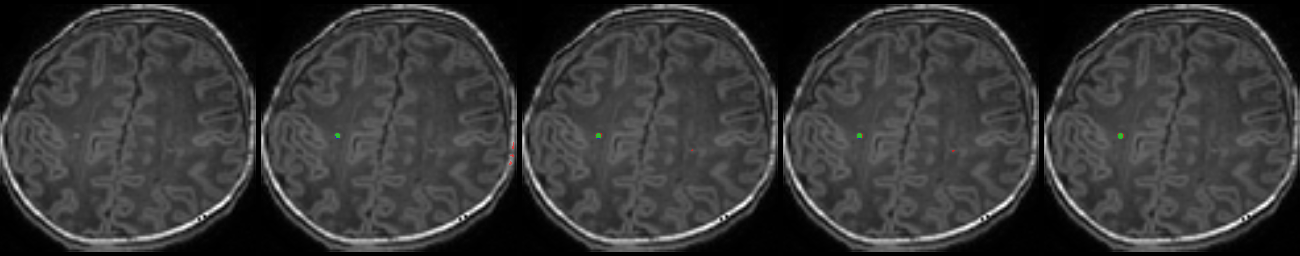 |
| 4 | 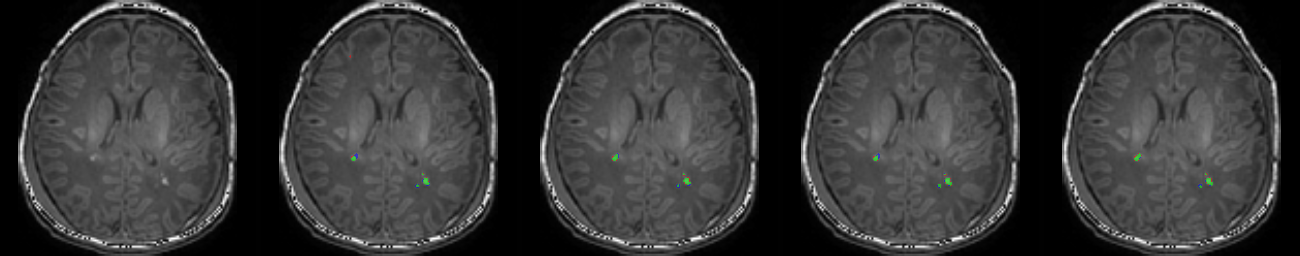 |
| 5 | 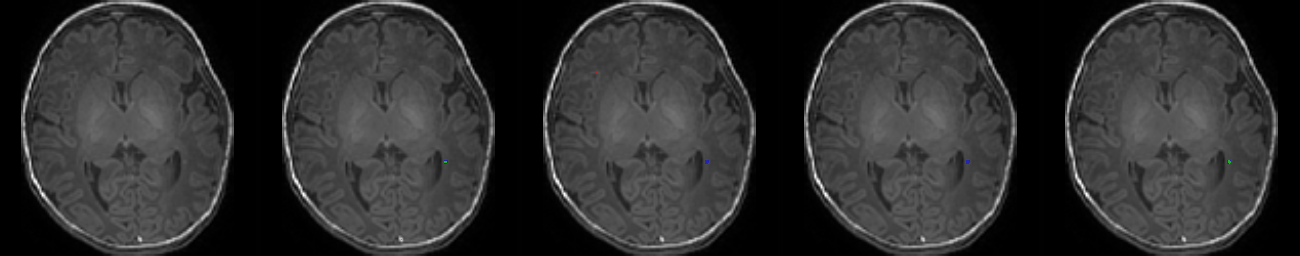 |
If you use Trident Segmentation CNN in your research, please cite the paper (Arxiv)
Liu, Yalong, Jie Li, Miaomiao Wang, Zhicheng Jiao, Jian Yang, and Xianjun Li. "Trident Segmentation CNN: A Spatiotemporal Transformation CNN for Punctate White Matter Lesions Segmentation in Preterm Neonates." arXiv preprint arXiv:1910.09773 (2019).