Go to KDD'23 Tutorial Link https://sunlabuiuc.github.io/PyHealth/
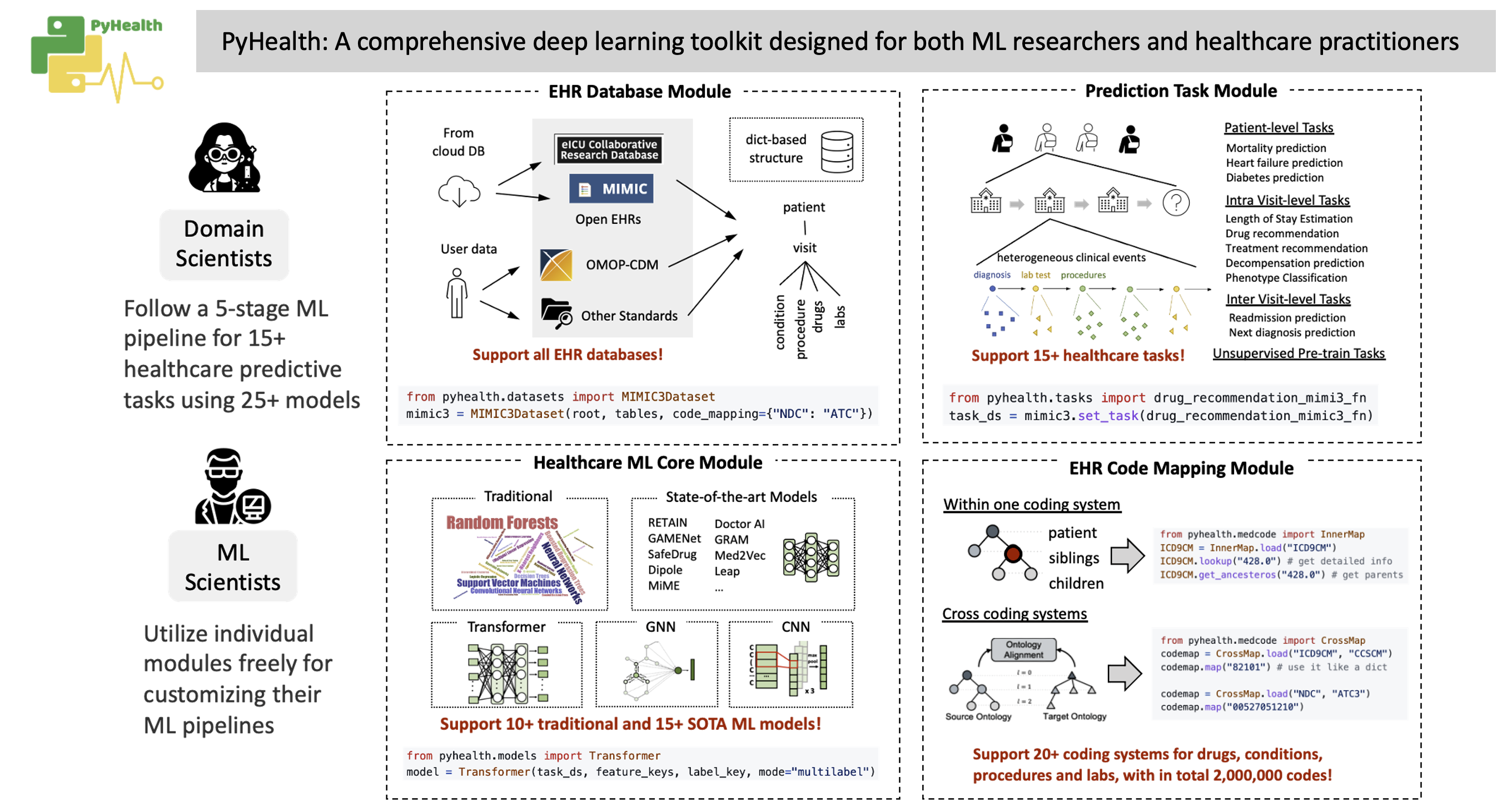
PyHealth is designed for both ML researchers and medical practitioners. We can make your healthcare AI applications easier to deploy and more flexible and customizable. [Tutorials]
[News!] Our PyHealth is accepted by KDD 2023 Tutorial Track! We will present a 3-hour tutorial on PyHealth at [KDD 2023], August 6-10, Long Beach, CA. The [hands-on tutorial jupyters] are available. Please check them out!
- You could install from PyPi:
pip install pyhealth- or from github source:
pip install .pyhealth provides these functionalities (we are still enriching some modules):
You can use the following functions independently:
- Dataset:
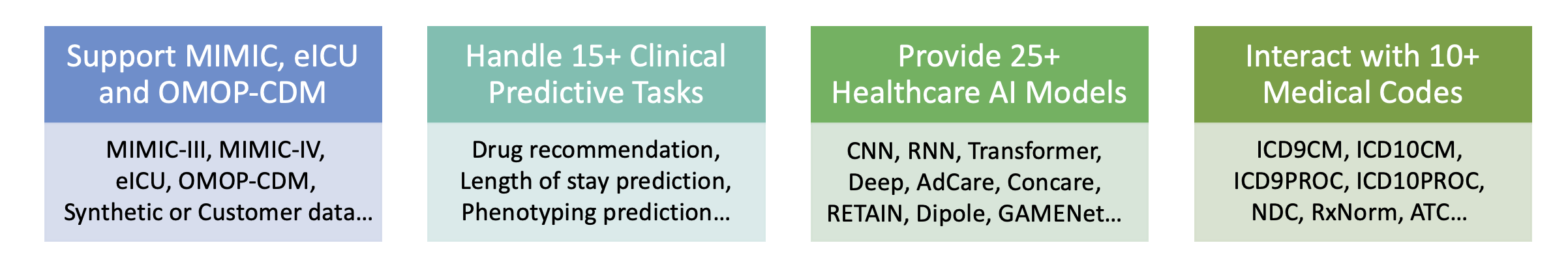
MIMIC-III,MIMIC-IV,eICU,OMOP-CDM,customized EHR datasets, etc. - Tasks:
diagnosis-based drug recommendation,patient hospitalization and mortality prediction,length stay forecasting, etc. - ML models:
CNN,LSTM,GRU,LSTM,RETAIN,SafeDrug,Deepr, etc.
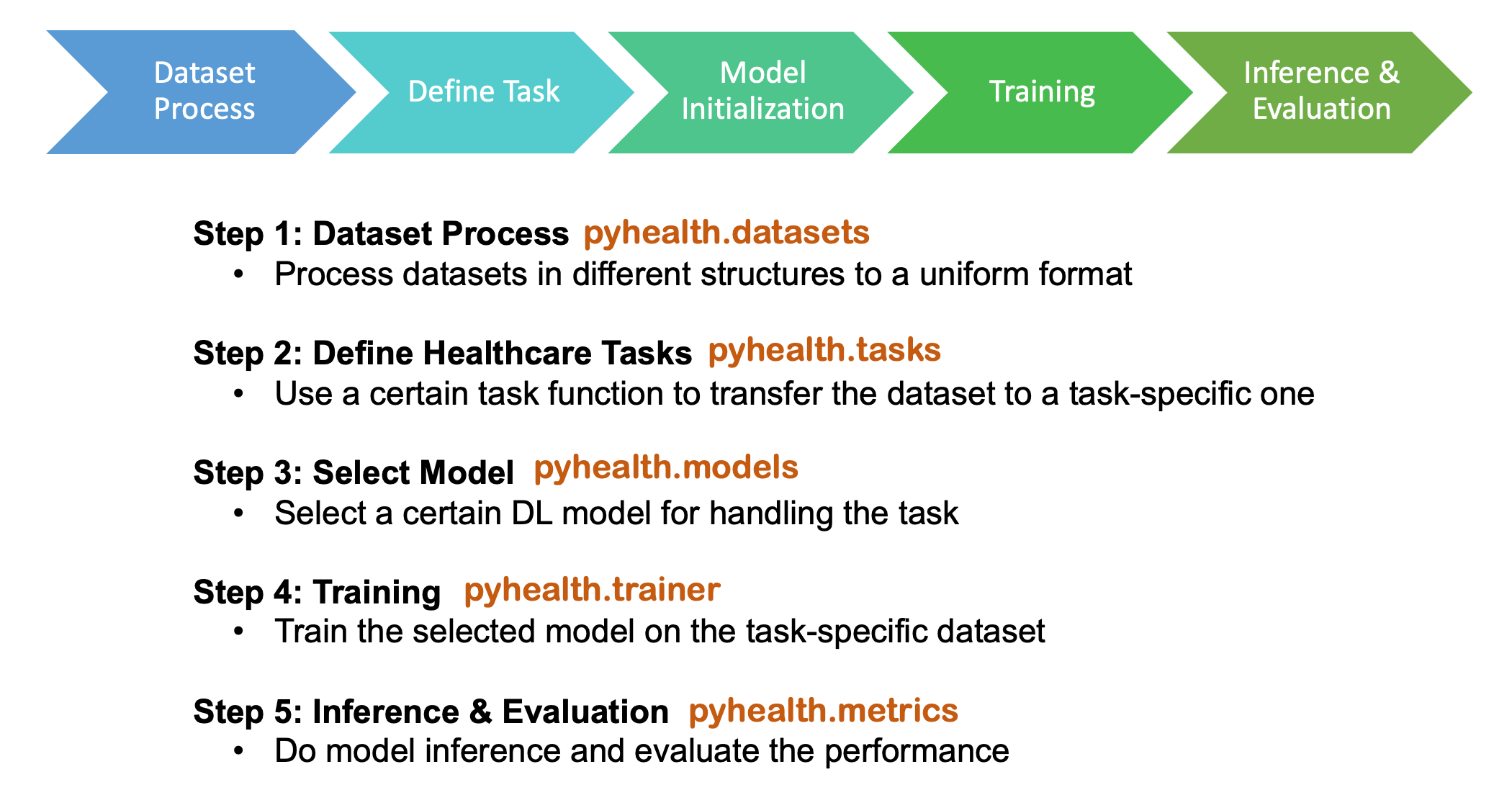
Building a healthcare AI pipeline can be as short as 10 lines of code in PyHealth.
All healthcare tasks in our package follow a five-stage pipeline:
We try hard to make sure each stage is as separate as possible, so that people can customize their own pipeline by only using our data processing steps or the ML models.
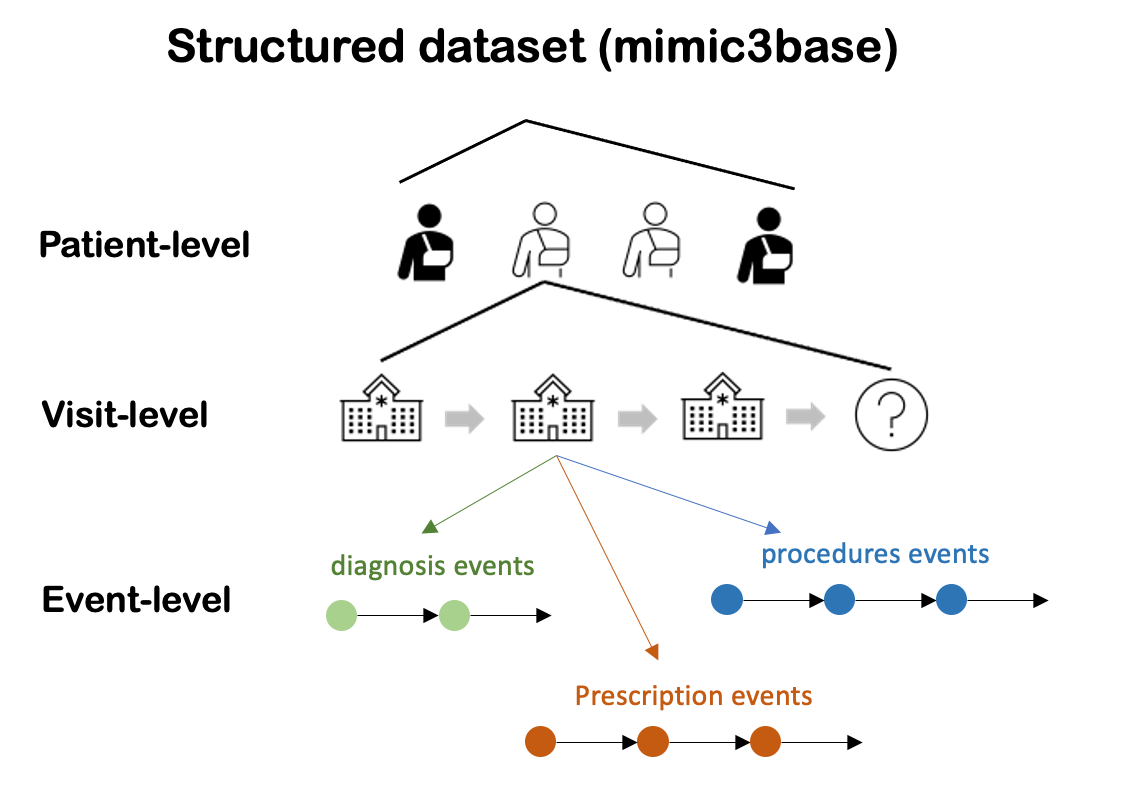
pyhealth.datasets provides a clean structure for the dataset, independent from the tasks. We support MIMIC-III, MIMIC-IV and eICU, etc. The output (mimic3base) is a multi-level dictionary structure (see illustration below).
from pyhealth.datasets import MIMIC3Dataset
mimic3base = MIMIC3Dataset(
# root directory of the dataset
root="https://storage.googleapis.com/pyhealth/Synthetic_MIMIC-III/",
# raw CSV table name
tables=["DIAGNOSES_ICD", "PROCEDURES_ICD", "PRESCRIPTIONS"],
# map all NDC codes to CCS codes in these tables
code_mapping={"NDC": "CCSCM"},
)pyhealth.tasks defines how to process each patient's data into a set of samples for the tasks. In the package, we provide several task examples, such as drug recommendation and length of stay prediction. It is easy to customize your own tasks following our template.
from pyhealth.tasks import readmission_prediction_mimic3_fn
mimic3sample = mimic3base.set_task(task_fn=readmission_prediction_mimic3_fn) # use default task
mimic3sample.samples[0] # show the information of the first sample
"""
{
'visit_id': '100183',
'patient_id': '175',
'conditions': ['5990', '4280', '2851', '4240', '2749', '9982', 'E8499', '42831', '34600'],
'procedures': ['0040', '3931', '7769'],
'drugs': ['N06DA02', 'V06DC01', 'B01AB01', 'A06AA02', 'R03AC02', 'H03AA01', 'J01FA09'],
'label': 0
}
"""
from pyhealth.datasets import split_by_patient, get_dataloader
train_ds, val_ds, test_ds = split_by_patient(mimic3sample, [0.8, 0.1, 0.1])
train_loader = get_dataloader(train_ds, batch_size=32, shuffle=True)
val_loader = get_dataloader(val_ds, batch_size=32, shuffle=False)
test_loader = get_dataloader(test_ds, batch_size=32, shuffle=False)pyhealth.models provides different ML models with very similar argument configs.
from pyhealth.models import Transformer
model = Transformer(
dataset=mimic3sample,
feature_keys=["conditions", "procedures", "drug"],
label_key="label",
mode="binary",
)pyhealth.trainer can specify training arguments, such as epochs, optimizer, learning rate, etc. The trainer will automatically save the best model and output the path in the end.
from pyhealth.trainer import Trainer
trainer = Trainer(model=model)
trainer.train(
train_dataloader=train_loader,
val_dataloader=val_loader,
epochs=50,
monitor="pr_auc_samples",
)pyhealth.metrics provides several common evaluation metrics (refer to Doc and see what are available).
# method 1
trainer.evaluate(test_loader)
# method 2
from pyhealth.metrics.binary import binary_metrics_fn
y_true, y_prob, loss = trainer.inference(test_loader)
binary_metrics_fn(y_true, y_prob, metrics=["pr_auc", "roc_auc"])pyhealth.codemap provides two core functionalities. This module can be used independently.
- For code ontology lookup within one medical coding system (e.g., name, category, sub-concept);
from pyhealth.medcode import InnerMap
icd9cm = InnerMap.load("ICD9CM")
icd9cm.lookup("428.0")
# `Congestive heart failure, unspecified`
icd9cm.get_ancestors("428.0")
# ['428', '420-429.99', '390-459.99', '001-999.99']
atc = InnerMap.load("ATC")
atc.lookup("M01AE51")
# `ibuprofen, combinations`
atc.lookup("M01AE51", "drugbank_id")
# `DB01050`
atc.lookup("M01AE51", "description")
# Ibuprofen is a non-steroidal anti-inflammatory drug (NSAID) derived ...
atc.lookup("M01AE51", "indication")
# Ibuprofen is the most commonly used and prescribed NSAID. It is very common over the ...- For code mapping between two coding systems (e.g., ICD9CM to CCSCM).
from pyhealth.medcode import CrossMap
codemap = CrossMap.load("ICD9CM", "CCSCM")
codemap.map("428.0")
# ['108']
codemap = CrossMap.load("NDC", "RxNorm")
codemap.map("50580049698")
# ['209387']
codemap = CrossMap.load("NDC", "ATC")
codemap.map("50090539100")
# ['A10AC04', 'A10AD04', 'A10AB04']pyhealth.tokenizer is used for transformations between string-based tokens and integer-based indices, based on the overall token space. We provide flexible functions to tokenize 1D, 2D and 3D lists. This module can be used independently.
from pyhealth.tokenizer import Tokenizer
# Example: we use a list of ATC3 code as the token
token_space = ['A01A', 'A02A', 'A02B', 'A02X', 'A03A', 'A03B', 'A03C', 'A03D', \
'A03F', 'A04A', 'A05A', 'A05B', 'A05C', 'A06A', 'A07A', 'A07B', 'A07C', \
'A12B', 'A12C', 'A13A', 'A14A', 'A14B', 'A16A']
tokenizer = Tokenizer(tokens=token_space, special_tokens=["<pad>", "<unk>"])
# 2d encode
tokens = [['A03C', 'A03D', 'A03E', 'A03F'], ['A04A', 'B035', 'C129']]
indices = tokenizer.batch_encode_2d(tokens)
# [[8, 9, 10, 11], [12, 1, 1, 0]]
# 2d decode
indices = [[8, 9, 10, 11], [12, 1, 1, 0]]
tokens = tokenizer.batch_decode_2d(indices)
# [['A03C', 'A03D', 'A03E', 'A03F'], ['A04A', '<unk>', '<unk>']]
# 3d encode
tokens = [[['A03C', 'A03D', 'A03E', 'A03F'], ['A08A', 'A09A']], \
[['A04A', 'B035', 'C129']]]
indices = tokenizer.batch_encode_3d(tokens)
# [[[8, 9, 10, 11], [24, 25, 0, 0]], [[12, 1, 1, 0], [0, 0, 0, 0]]]
# 3d decode
indices = [[[8, 9, 10, 11], [24, 25, 0, 0]], \
[[12, 1, 1, 0], [0, 0, 0, 0]]]
tokens = tokenizer.batch_decode_3d(indices)
# [[['A03C', 'A03D', 'A03E', 'A03F'], ['A08A', 'A09A']], [['A04A', '<unk>', '<unk>']]]We provide the following tutorials to help users get started with our pyhealth.
Tutorial 0: Introduction to pyhealth.data [Video]
Tutorial 1: Introduction to pyhealth.datasets [Video]
Tutorial 2: Introduction to pyhealth.tasks [Video]
Tutorial 3: Introduction to pyhealth.models [Video]
Tutorial 4: Introduction to pyhealth.trainer [Video]
Tutorial 5: Introduction to pyhealth.metrics [Video]
Tutorial 6: Introduction to pyhealth.tokenizer [Video]
Tutorial 7: Introduction to pyhealth.medcode [Video]
The following tutorials will help users build their own task pipelines.
Pipeline 1: Drug Recommendation [Video]
Pipeline 2: Length of Stay Prediction [Video]
Pipeline 3: Readmission Prediction [Video]
Pipeline 4: Mortality Prediction [Video]
Pipeline 5: Sleep Staging [Video]
We provided the advanced tutorials for supporting various needs.
Advanced Tutorial 1: Fit your dataset into our pipeline [Video]
Advanced Tutorial 2: Define your own healthcare task
Advanced Tutorial 3: Adopt customized model into pyhealth [Video]
Advanced Tutorial 4: Load your own processed data into pyhealth and try out our ML models [Video]
We provide the processing files for the following open EHR datasets:
| Dataset | Module | Year | Information |
|---|---|---|---|
| MIMIC-III | pyhealth.datasets.MIMIC3Dataset |
2016 | MIMIC-III Clinical Database |
| MIMIC-IV | pyhealth.datasets.MIMIC4Dataset |
2020 | MIMIC-IV Clinical Database |
| eICU | pyhealth.datasets.eICUDataset |
2018 | eICU Collaborative Research Database |
| OMOP | pyhealth.datasets.OMOPDataset |
OMOP-CDM schema based dataset | |
| SleepEDF | pyhealth.datasets.SleepEDFDataset |
2018 | Sleep-EDF dataset |
| SHHS | pyhealth.datasets.SHHSDataset |
2016 | Sleep Heart Health Study dataset |
| ISRUC | pyhealth.datasets.ISRUCDataset |
2016 | ISRUC-SLEEP dataset |
| Model Name | Type | Module | Year | Summary | Reference |
|---|---|---|---|---|---|
| Multi-layer Perceptron | deep learning | pyhealth.models.MLP |
1986 | MLP treats each feature as static | Backpropagation: theory, architectures, and applications |
| Convolutional Neural Network (CNN) | deep learning | pyhealth.models.CNN |
1989 | CNN runs on the conceptual patient-by-visit grids | Handwritten Digit Recognition with a Back-Propagation Network |
| Recurrent Neural Nets (RNN) | deep Learning | pyhealth.models.RNN |
2011 | RNN (includes LSTM and GRU) can run on any sequential level (e.g., visit by visit sequences) | Recurrent neural network based language model |
| Transformer | deep Learning | pyhealth.models.Transformer |
2017 | Transformer can run on any sequential level (e.g., visit by visit sequences) | Atention is All you Need |
| RETAIN | deep Learning | pyhealth.models.RETAIN |
2016 | RETAIN uses two RNN to learn patient embeddings while providing feature-level and visit-level importance. | RETAIN: An Interpretable Predictive Model for Healthcare using Reverse Time Attention Mechanism |
| GAMENet | deep Learning | pyhealth.models.GAMENet |
2019 | GAMENet uses memory networks, used only for drug recommendation task | GAMENet: Graph Attention Mechanism for Explainable Electronic Health Record Prediction |
| MICRON | deep Learning | pyhealth.models.MICRON |
2021 | MICRON predicts the future drug combination by instead predicting the changes w.r.t. the current combination, used only for drug recommendation task | Change Matters: Medication Change Prediction with Recurrent Residual Networks |
| SafeDrug | deep Learning | pyhealth.models.SafeDrug |
2021 | SafeDrug encodes drug molecule structures by graph neural networks, used only for drug recommendation task | SafeDrug: Dual Molecular Graph Encoders for Recommending Effective and Safe Drug Combinations |
| MoleRec | deep Learning | pyhealth.models.MoleRec |
2023 | MoleRec encodes drug molecule in a substructure level as well as the patient's information into a drug combination representation, used only for drug recommendation task | MoleRec: Combinatorial Drug Recommendation with Substructure-Aware Molecular Representation Learning |
| Deepr | deep Learning | pyhealth.models.Deepr |
2017 | Deepr is based on 1D CNN. General purpose. | Deepr : A Convolutional Net for Medical Records |
| ContraWR Encoder (STFT+CNN) | deep Learning | pyhealth.models.ContraWR |
2021 | ContraWR encoder uses short time Fourier transform (STFT) + 2D CNN, used for biosignal learning | Self-supervised EEG Representation Learning for Automatic Sleep Staging |
| SparcNet (1D CNN) | deep Learning | pyhealth.models.SparcNet |
2023 | SparcNet is based on 1D CNN, used for biosignal learning | Development of Expert-level Classification of Seizures and Rhythmic and Periodic Patterns During EEG Interpretation |
| TCN | deep learning | pyhealth.models.TCN |
2018 | TCN is based on dilated 1D CNN. General purpose | Temporal Convolutional Networks |
| AdaCare | deep learning | pyhealth.models.AdaCare |
2020 | AdaCare uses CNNs with dilated filters to learn enriched patient embedding. It uses feature calibration module to provide the feature-level and visit-level interpretability | AdaCare: Explainable Clinical Health Status Representation Learning via Scale-Adaptive Feature Extraction and Recalibration |
| ConCare | deep learning | pyhealth.models.ConCare |
2020 | ConCare uses transformers to learn patient embedding and calculate inter-feature correlations. | ConCare: Personalized Clinical Feature Embedding via Capturing the Healthcare Context |
| StageNet | deep learning | pyhealth.models.StageNet |
2020 | StageNet uses stage-aware LSTM to conduct clinical predictive tasks while learning patient disease progression stage change unsupervisedly | StageNet: Stage-Aware Neural Networks for Health Risk Prediction |
| Dr. Agent | deep learning | pyhealth.models.Agent |
2020 | Dr. Agent uses two reinforcement learning agents to learn patient embeddings by mimicking clinical second opinions | Dr. Agent: Clinical predictive model via mimicked second opinions |
| GRASP | deep learning | pyhealth.models.GRASP |
2021 | GRASP uses graph neural network to identify latent patient clusters and uses the clustering information to learn patient | GRASP: Generic Framework for Health Status Representation Learning Based on Incorporating Knowledge from Similar Patients |
@inproceedings{pyhealth2023yang,
author = {Yang, Chaoqi and Wu, Zhenbang and Jiang, Patrick and Lin, Zhen and Gao, Junyi and Danek, Benjamin and Sun, Jimeng},
title = {{PyHealth}: A Deep Learning Toolkit for Healthcare Predictive Modeling},
url = {https://github.com/sunlabuiuc/PyHealth},
booktitle = {Proceedings of the 27th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD) 2023},
year = {2023}
}