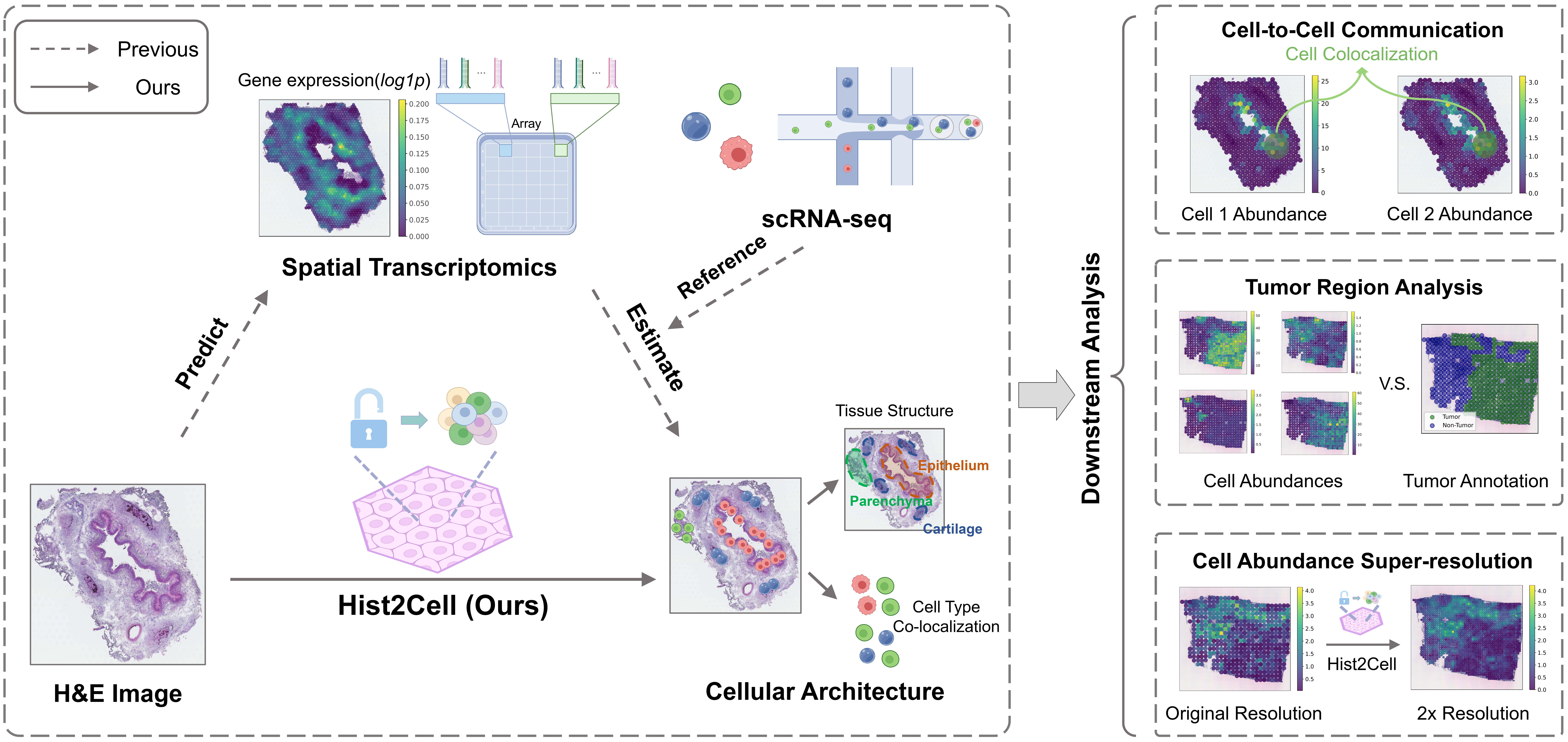
Hist2Cell is a Vision Graph-Transformer framework that accurately predicts fine-grained cell type abundances directly from histology images. It facilitates cost-efficient, high-resolution cellular mapping of tissues, significantly advancing spatial biology studies and clinical diagnostics.
For more details about this stuy, please check our paper Hist2Cell: Deciphering Fine-grained Cellular Architectures from Histology Images.
Create the environment with conda commands:
conda create -n Hist2Cell python=3.11
conda activate Hist2Cell
Install the dependencies:
git clone https://github.com/Weiqin-Zhao/Hist2Cell.git
cd Hist2Cell
pip install -r requirements.txt
pip install torch==2.0.1 torchvision==0.15.2 torchaudio==2.0.2 --index-url https://download.pytorch.org/whl/cu118
pip install torch_geometric
pip install pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.0.0+cu118.html
All datasets are previously published and publicly accessible.
- The healthy lung dataset was downloaded from https://5locationslung.cellgeni.sanger.ac.uk/.
- The her2st dataset was obtained from https://github.com/almaan/her2st.
- The STNet dataset was sourced from https://data.mendeley.com/datasets/29ntw7sh4r/5.
- The TCGA dataset was acquired from the Genomic Data Commons Data Portal at https://portal.gdc.cancer.gov/.
- The scRNA-seq data from the Human Breast Cell Atlas (HBCA) was downloaded from CELLxGENE at https://cellxgene.cziscience.com/collections/4195ab4c-20bd-4cd3-8b3d-65601277e731.
We also provide example raw data in ./example_raw_data and the pre-process tutorials in ./data_preparation_tutorial.ipynb. Users can pre-process their own datasets following the same steps for inference/training/fine-tuning.
We provide processed example data of the healthy lung dataset in ./example_data/humanlung_cell2location and ./example_data/humanlung_cell2location_2x (for super-resolved cell abundances usage).
We upload the data in compressed format via Onedrive, please download the data and unzip them using tar -xzvf command.
The processed data of breast cancer will be realsed soon.
We have uploaded the checkpoint weight for healthy lung dataset in ./model_weights.
For training on your own dataset, we provide detailed training tutorials in ./tutorial_training/training_tutorial.ipynb with the example data we uploaded.
After preparing your own dataset following ./tutorial_data_preparation/data_preparation_tutorial.ipynb, users can train/finetune Hist2Cell on their own dataset for further cellular analysis.
We uploaded the pretrained model weights on healthy human lung dataset in ./model_weights and provide detailed tutorial steps for the cellular analysis conducted in our study:
./tutorial_analysis_evaluation/cell_abundance_visulization_tutorial.ipynb: visualizeHist2Cellpredicted fine-grained cell abundance for biological finding validatoin, in this tutorial, we generate the figures used inFig 2.fandFig 3.bcin our paper;./tutorial_analysis_evaluation/key_cell_evaluation_tutorial.ipynb: evalute the prediction performance ofHist2Cellon serveral key cell types of interest, in this tutorial, we generate the figures used inFig 2.d;./tutorial_analysis_evaluation/cell_colocalization_tutorial.ipynb: analyse the cell co-localization patterns from histology image usingHist2Cell, in this tutorial, we generate the figures used inFig 2.f;./tutorial_analysis_evaluation/super_resovled_cell_abundance_tutorial.ipynb: produce super-resolved fine-grained cell type abundances usingHist2Cellfor biological reserach, in this tutorial, we generate the figures used inFig 6.b.
If you find our paper/code/results useful, please consider cite us using the following BibTex entry.
@article{zhao2024hist2cell,
title={Hist2Cell: Deciphering Fine-grained Cellular Architectures from Histology Images},
author={Zhao, Weiqin and Liang, Zhuo and Huang, Xianjie and Huang, Yuanhua and Yu, Lequan},
journal={bioRxiv},
pages={2024--02},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}