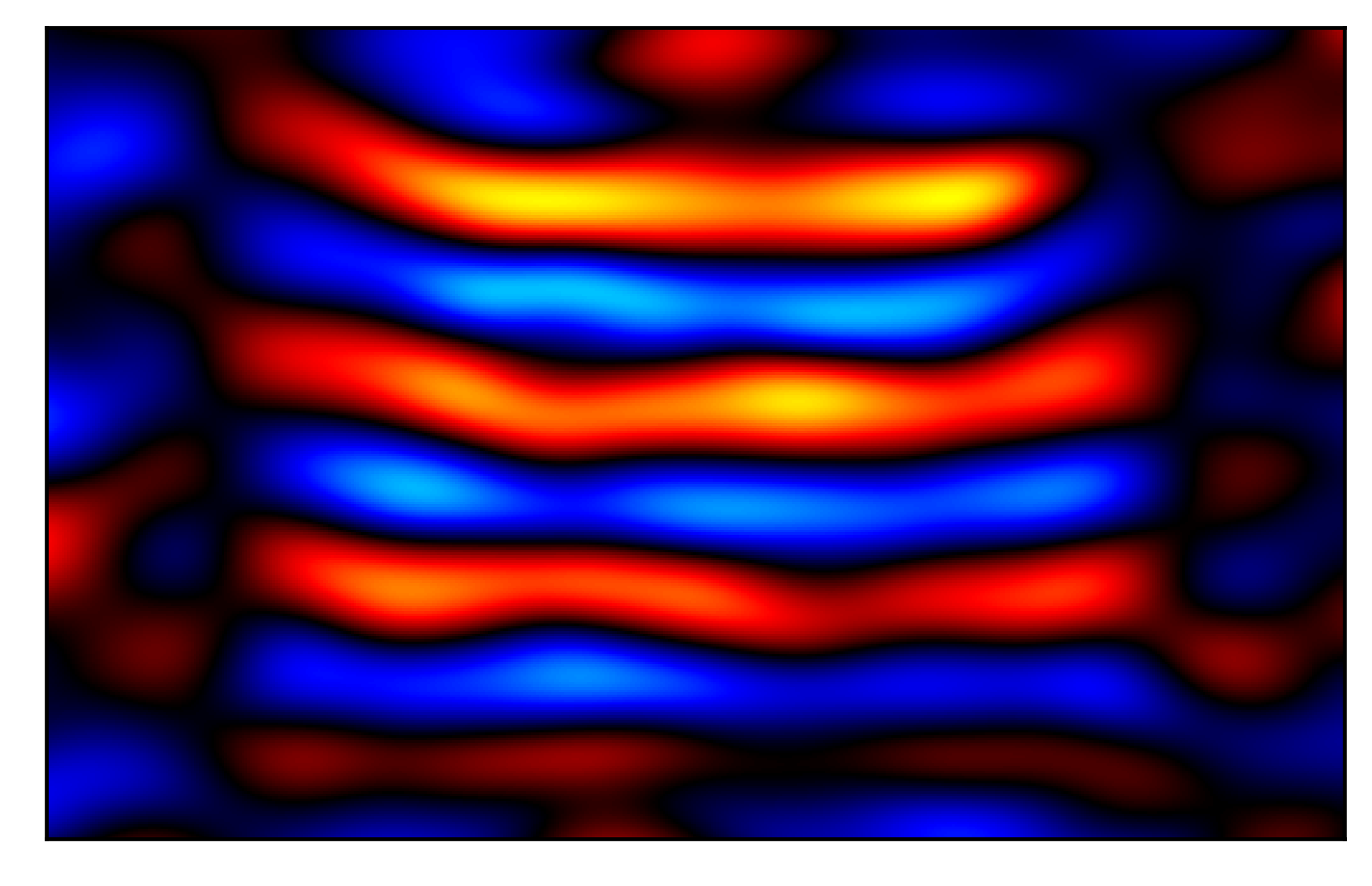
The goal of this project is to perform elasticity reconstruction for magnetic resonance elastography using sinusoidal representation networks (SIRENs).
This image shows the continuous representation of a shear wave image learned using SIREN, which enables super-resolution and solving of partial differental equations.
conda env create -n MRE-SIREN --file environment.yml
conda activate MRE-SIREN
python -m ipykernel install --user --name=MRE-SIREN
mre_siren/: Python code modulebioqic.py: Code for working with BIOQIC data setmodels.py: Torch implementation of SIRENpde.py: Torch differential operatorsphase.py: Numpy wave image preprocessing and MDEV
notebooks/: Jupyter notebooksBIOQIC-data-exploration.ipynb: Loading and visualizing the BIOQIC data setMDEV-inversion-method.ipynb: Development of phase preprocessing and MDEV inversion in PythonSIREN-testing.ipynb: Initial toy implementation of SIREN model on random dataMRE-SIREN-training.ipynb: Training SIREN on the BIOQIC data set and performing inversionMRE-SIREN-optimization.ipynb: SIREN hyperparameter search for elasticity reconstructionMRE-SIREN-evaluation.ipynb: Ground truth evaluation of SIREN compared to MDEV
download_data.sh: Script to download BIOQIC data setenvironment.yml: Conda environment filetrain.py: Main training script