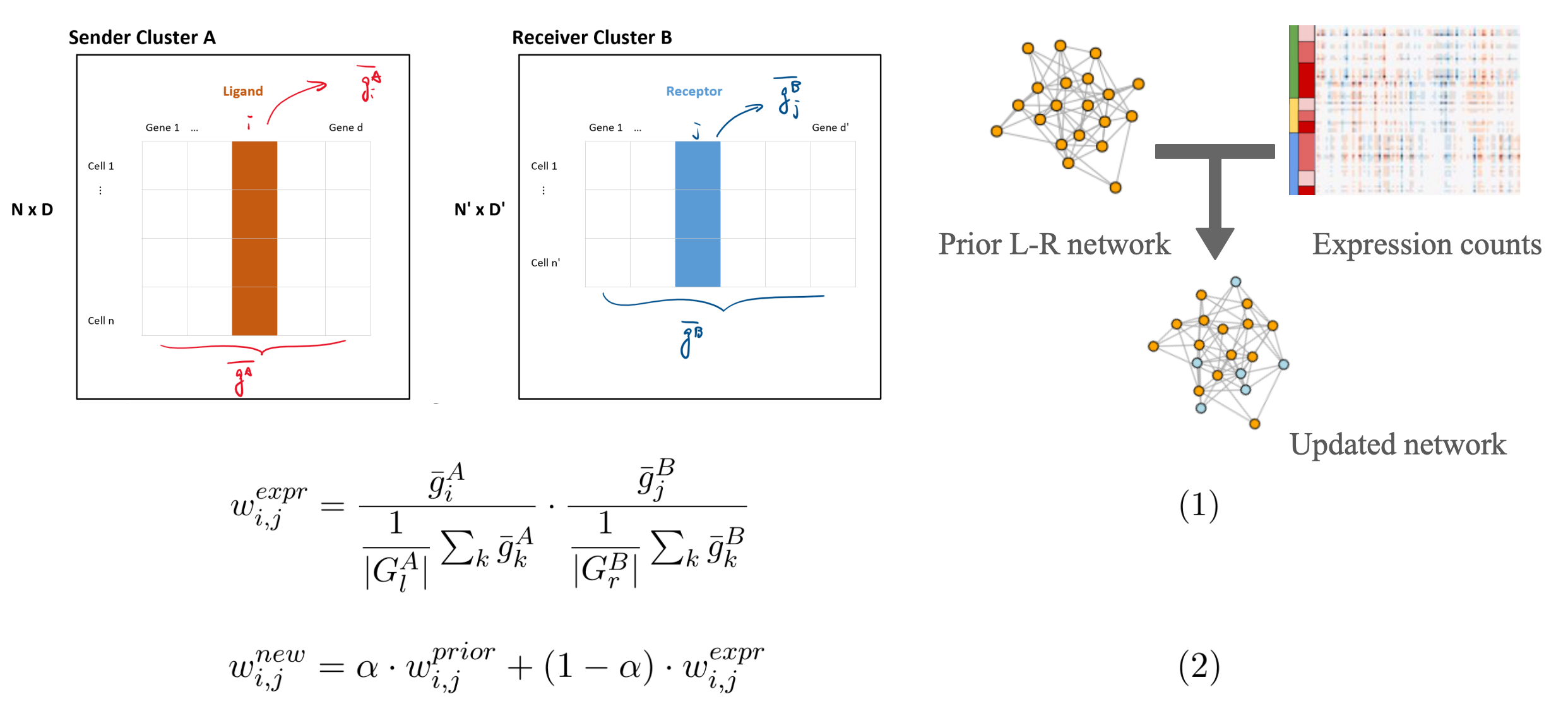
We applied NicheNet to infer intercellular Ligand-Receptor (L-R) interactions from scRNA-seq data. In particular, we investigated the mutual interactions between cancer stem cells (CSCs) and tumor associated macrophages (TAMs) retrieved from the following dataset via NicheNet. We also modified the L-R interaction weights by combining NicheNet's prior network scores and our updated scores from the dataset-specific expression values.
Python: Numpy, Pandas, Scanpy
R: nichenetr
Installation of NicheNet from their github page:
Installation typically takes a few minutes, depending on the number of dependencies that has already been installed on your pc. You can install nichenetr (and required dependencies) from github with:
install.packages("devtools") devtools::install_github("saeyslab/nichenetr")nichenetr was tested on both Windows and Linux (most recently tested R version: R 4.0.0)
Scripts
preprocessing.ipynbPreprocessing, data cleaning, clustering, identification of expressed genes & marker genes for each clusterweight_calculatioo.ipypbUpdate the edge scores of the L-R prior network from NicheNet with expression values (see the concept figure)rl_utils.RUtility & visualization functions for ligand receptor interaction inferencerl_predictions.RmdL-R inference with NicheNet's prior networkrl_predictions_weighted.RmdL-R inference with our updated networkvisualize.RmdVisualization of top L-R interactionsBrain.RDifferential analysis & gene ontology, pathway analysis
Directories
- dataset: Original & preprocessed count matrices
- supplementary_materials: supplementary table for final report
- plots: Output plots with NicheNet's prior network
- plots_weighted: Output plots with our updated network
- top_rl_pairs: Top inferreed L-R pairs with NicheNet's prior network
- top_rl_pairs_weighted: Top inferred L-R pairs with our updated network