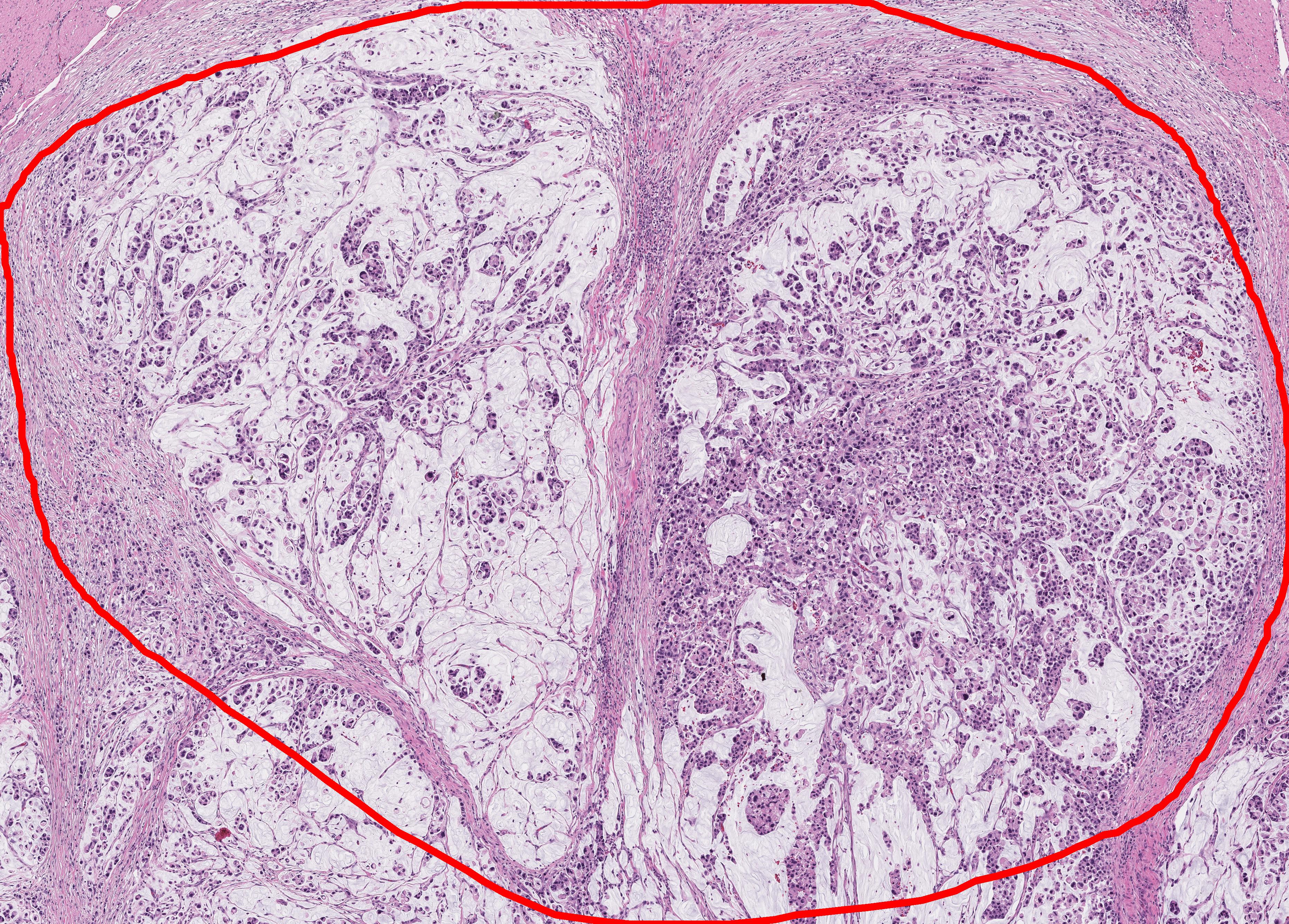
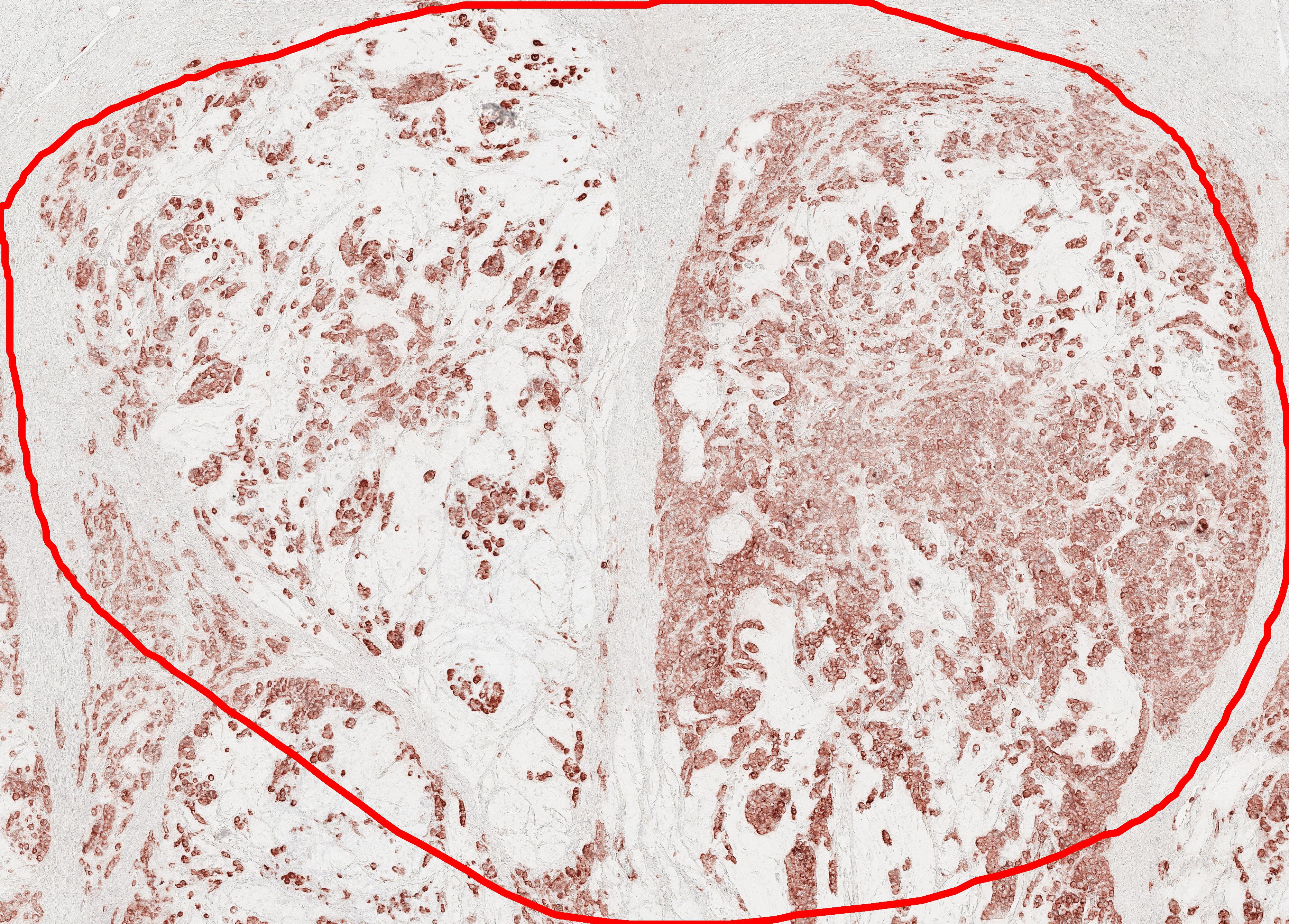
This repository is for virtual staining (transforming H&E image into CK (cytokeratin) image).
- python >= 3.6
- numpy >=1.17.4
- matplotlib >=3.1.1
- opencv-python >= 4.1.2.30
- openslide-python >= 1.1.2
- pandas >= 1.1.3
- scikit-image >= 0.15.0
- scikit-learn >= 0.23.2
- tifffile >= 2020.7.4
- torch >= 1.5.1 (https://pytorch.org/)
- torchvision >= 0.6.1
- tqdm >= 4.50.2
- openslide >= 3.4.1 (https://openslide.org/)
- -input_path: path for input H&E image (.svs). example images in
data/input - -output_dir: directory for output CK image
- -model_checkpoint: path for model weight (https://drive.google.com/file/d/1-1TqdXRCApmqjSXXiDNwsMbo4Uz8EITJ/view?usp=sharing)
python run_ck_virtual_staining.py -input_path data/input/example.svs -output_dir data/output -model_checkpoint data/checkpoint/model.pth
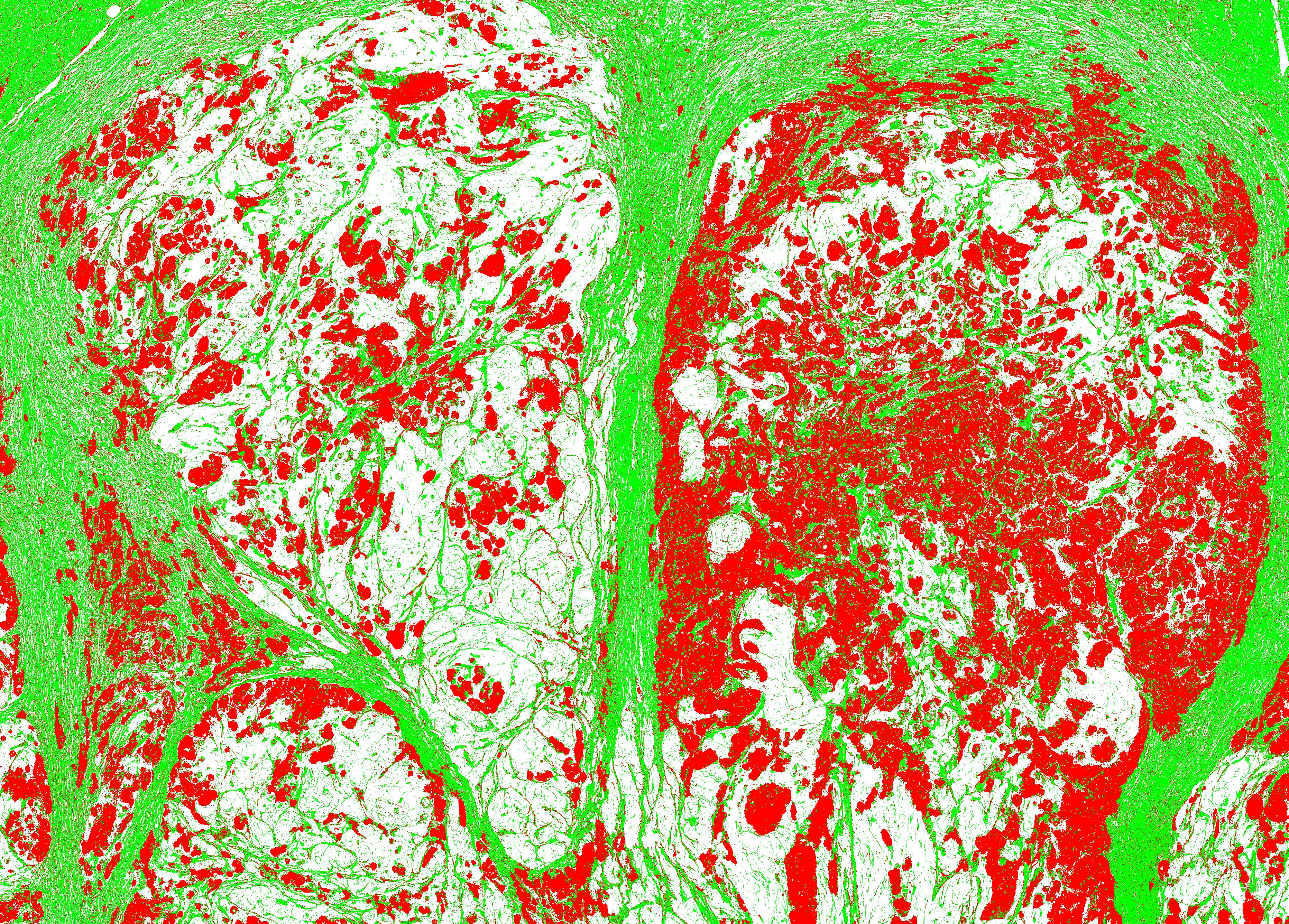
| Input | Output_CK | Output_Segmentation (Tumor(red), Stroma(green)) |
|---|---|---|
 |
 |
 |
Hong, Y., Heo, Y.J., Kim, B. et al. Deep learning-based virtual cytokeratin staining of gastric carcinomas to measure tumor–stroma ratio. Sci Rep 11, 19255 (2021). https://doi.org/10.1038/s41598-021-98857-1