UniMiSS-code
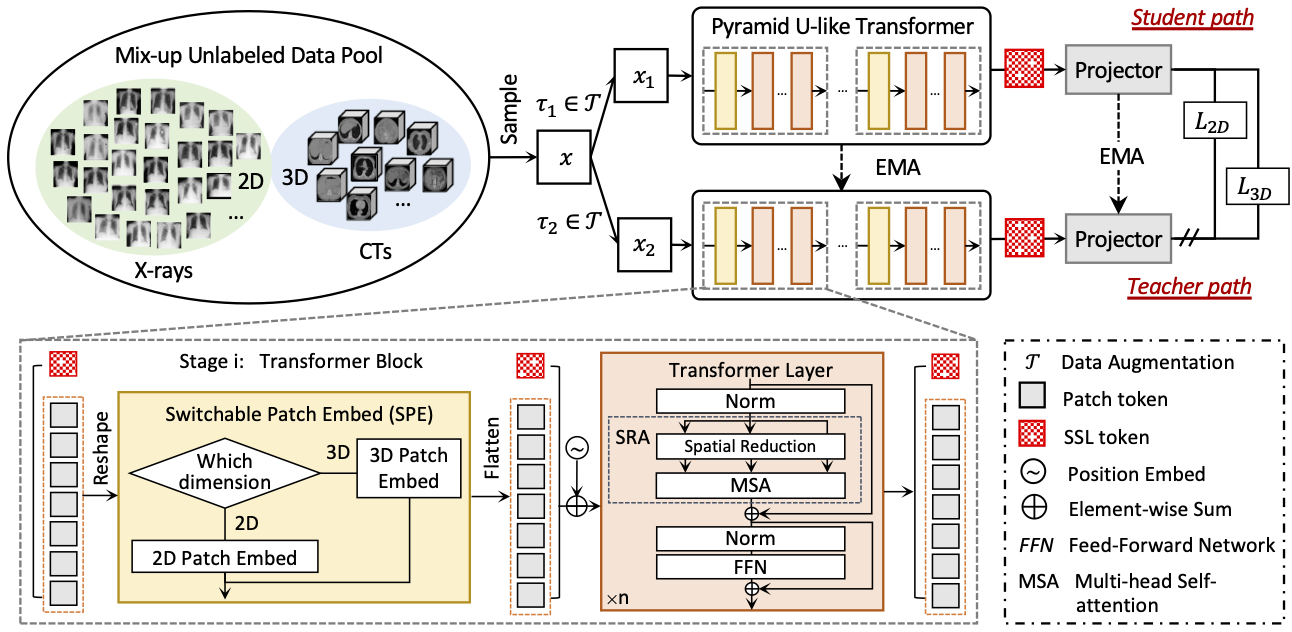
This is the official pytorch implementation of our ECCV 2022 paper "UniMiSS: Universal Medical Self-Supervised Learning via Breaking Dimensionality Barrier". In this paper, we advocate bringing a wealth of 2D images like chest X-rays as compensation for the lack of 3D data, aiming to build a universal medical self-supervised representation learning framework, called UniMiSS. We conduct expensive experiments on six 3D/2D medical image analysis tasks, including segmentation and classification. The results show that the proposed UniMiSS achieves promising performance on various downstream tasks, outperforming the ImageNet pre-training and other advanced SSL counterparts substantially.
Requirements
CUDA 11.0
Python 3.7
Pytorch 1.7.1
Torchvision 0.8.2
Usage
Installation
- Clone this repo
git clone https://github.com/YtongXie/UniMiSS-code.git
cd UniMiSS-code
- Create virtual environment
conda create --name UniMiSS python=3.7
conda activate UniMiSS
- Install dependencies as below
pip install torch==1.7.1+cu110 torchvision==0.8.2+cu110 torchaudio==0.7.2 -f https://download.pytorch.org/whl/torch_stable.html
pip install batchgenerators==0.21
pip install dicom2nifti==2.3.0
pip install matplotlib==3.4.2
pip install ml-collections==0.1.0
pip install nibabel==3.2.1
pip install numpy==1.19.5
pip install opencv-python==4.5.2.52
pip install Pillow==8.2.0
pip install pydicom==2.1.2
pip install scikit-image==0.18.1
pip install scikit-learn==0.24.2
pip install scipy==1.6.3
pip install SimpleITK==1.2.4
pip install sklearn==0.0
pip install tensorboard==2.5.0
pip install torchio==0.18.39
pip install timm
Data Preparation
- cd UniMiSS/data
- Download MOTS data, LIDC-IDRI dataset, Tianchi dataset, RibFrac dataset, and TCIACT dataset, then put them into
3D images - Resample CT volumes to a unified voxel size of 1.0×1.0×3.0 mm3.
- Run
python extract_subvolumes.pyto extract about 120k sub-volumes, and put them into3D subvolumes - The image folder of 3D images should be like:
data/3D subvolumes/
├── LIDC
| ├── LKDS-00001_dep0.nii
| ├── ...
├── Tianchi
| ├── LIDC-IDRI-0001_dep0.nii
| ├── ...
├── RibFrac
| ├── RibFrac1-image_dep0.nii
| ├── ...
├── ...
- Download NIH ChestX-ray8 dataset
- Resize ChestX-ray8 images into 512×512 and put the resized images into
2D images. - The image folder of ChestX-ray8 should look like this:
data/2D images/
├── 00000001_000.png
├── 00000001_001.png
├── ...
Training
- cd UniMiSS
- Run
sh run.shfor self-supervised pre-training.
Pretrained weights
- Pretrained models are available in UniMiss_tiny and UniMiss_small.
Finetuning
- Finetuning codes are available in Downstream.
Citation
If this code is helpful for your study, please cite:
@article{UniMiSS,
title={UniMiSS: Universal Medical Self-Supervised Learning via Breaking Dimensionality Barrier},
author={Xie, Yutong and Zhang, Jianpeng and Xia, Yong and Wu, Qi},
booktitle={ECCV},
year={2022}
}
Acknowledgements
Part of codes is reused from the DINO. Thanks to Caron et al. for the codes of DINO.
Contact
Yutong Xie (yutong.xie678@gmail.com)