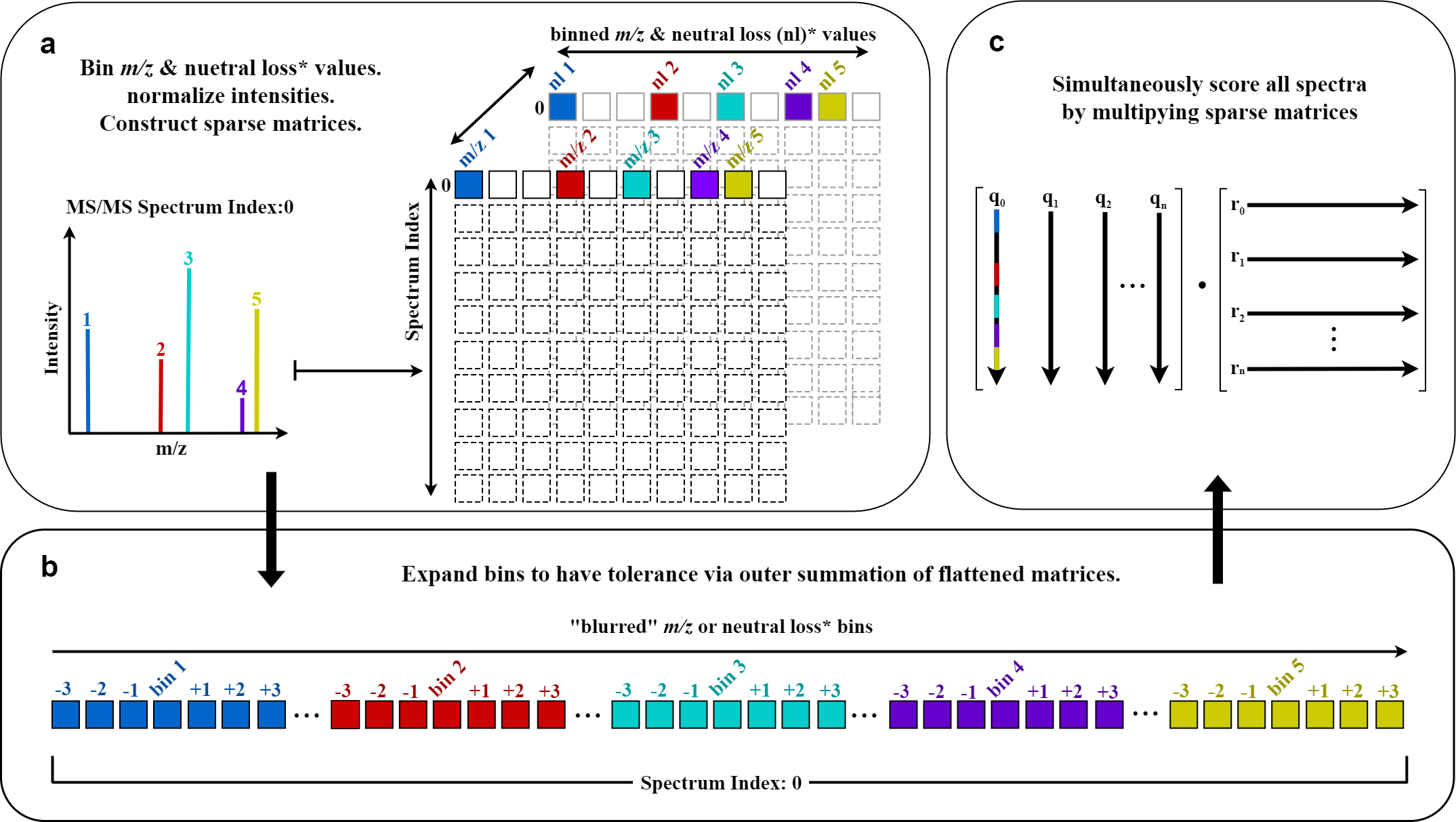
BLINK (Blur-and-Link) is a Python package for efficiently generating cosine-based similarity scores and matching ion counts for large numbers of fragmentation mass spectra.
Use the package manager conda to install environment.yml for all BLINK and example notebook requirements.
conda env create -f environment.yml- python3
- numpy
- scipy
- pandas
- pyteomics
- pymzml
- networkx
Please see for descriptions and usage of primary functions and all optional parameters
tutorial/blink_tutorial.ipynb
>> ./blink.py --help
usage: blink.py [-h] [--trim] [--dedup] [-b B] [-i I] [-t T] [-d [D ...]] [-r R] [-s S] [-m M] [--fast_format] [-f] [-o O] F [F ...]
BLINK discretizes mass spectra (given .mgf inputs), and scores discretized spectra (given .npz inputs)
positional arguments:
F files to process
optional arguments:
-h, --help show this help message and exit
--trim remove empty spectra when discretizing
--dedup deduplicate fragment ions within 2 times bin_width
-b B, --bin_width B width of bins in mz
-i I, --intensity_power I
power to raise intensites to in when scoring
-t T, --tolerance T maximum tolerance in mz for fragment ions to match
-d [D ...], --mass_diffs [D ...]
mass diffs to network
-r R, --react_steps R
recursively combine mass_diffs within number of reaction steps
-s S, --min_score S minimum score to include in output
-m M, --min_matches M
minimum matches to include in output
--fast_format use fast .npz format to store scores instead of .tab
-f, --force force file(s) to be remade if they exist
-o O, --out_dir O change output location for output file(s)
# Discretize fragmentation mass spectra to sparse matrix format (.npz)
# small = 1e2 spectra, medium = 1e4 spectra
>> blink.py ./example/small.mgf
small.npz
>> blink.py ./example/medium.mgf
medium.npz
# Compute all-by-all cosine scores and # matching ions for each fragmentation mass spectrum
>> blink.py ./example/small.npz
small.tab
# Compute A-vs-B cosine scores and # matching ions for each fragmentation mass spectrum
>> blink.py ./example/small.npz ./example/medium.npz
small_medium.tabPull requests are welcome.
For major changes, please open an issue first to discuss what you would like to change.