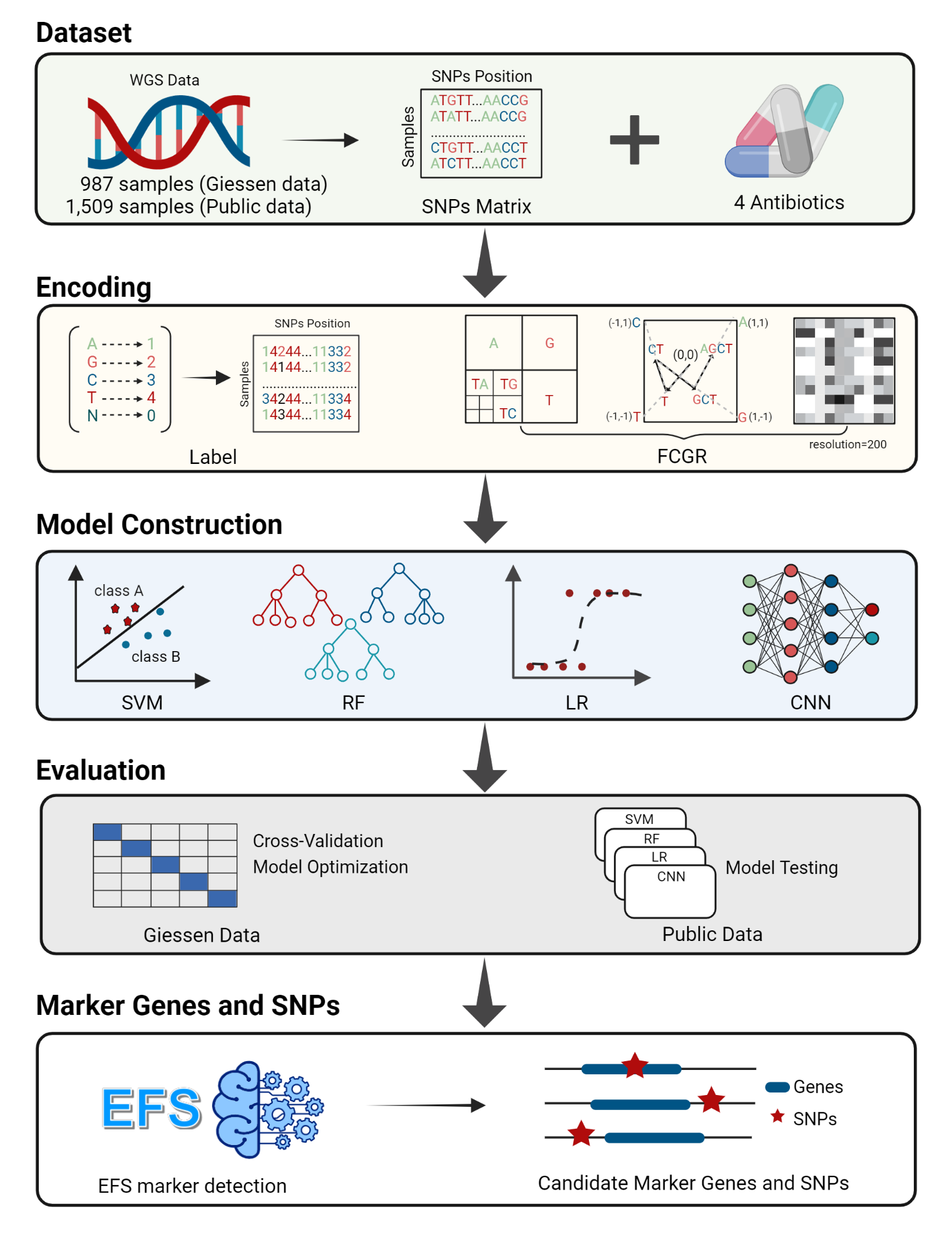
Antimicrobial resistance (AMR) is one of the biggest global problems threatening human and animal health. Rapid and accurate AMR diagnostic methods are thus very urgently needed. However, traditional antimicrobial susceptibility testing (AST) is time-consuming, low throughput, and viable only for cultivable bacteria. Machine learning methods may pave the way for automated AMR prediction based on genomic data of the bacteria. However, comparing different machine learning methods for the prediction of AMR based on different encodings and whole-genome sequencing data without previously known knowledge remains to be done.
In the current study, we evaluated logistic regression (LR), support vector machine (SVM), random forest (RF), and convolutional neural network (CNN) for the prediction of AMR for the antibiotics ciprofloxacin (CIP), cefotaxime (CTX), ceftazidime (CTZ), and gentamicin (GEN) based on WGS data with label encoding and FCGR encoding.
-
Variants Calling
- Here, we called variants using
bcftoolssoftware and E.coli K-12 strain. MG1655 as reference genome. You can also use other tools for variants calling.
- Here, we called variants using
-
SNP-matrix
-
We then extracted SNPs variants, reference alleles, and their positions, and merged all isolates based on the positions of reference alleles.
-
The final format of SNP-matrix (N replaces a locus without variation):
Sample_name Position_1 Position_2 Position_3 ... Position_n Ref_allele A T G ... C Sample_1 G A A ... T Sample_2 G N A ... T Sample_3 G A C ... G ... ... ... ... ... ... Sample_m T A A ... T
-
-
Encoding SNP-natrix
- Label encoding
- The A, G, C, T, N in the SNP_matrix were converted to 1, 2, 3, 4, and 0.
- One-hot encoding
- Each allele is encoded into a bianry matrix.
- Label encoding
Please refer to the format of "example_file.7z" to prepare your data and test the pipeline. We also provide the final SNPs matrix and phenotypic data for four drugs, which contains 809 E. coli strains "Giessen_dataset.zip". (The sample size is different from the one we used in the paper)