An R package to create “Table 1”, description of baseline characteristics
Creates “Table 1”, i.e., description of baseline patient characteristics, which is essential in every medical research. Supports both continuous and categorical variables, as well as p-values and standardized mean differences. Weighted data are supported via the survey package.
tableone was inspired by descriptive statistics functions in Deducer , a Java-based GUI package by Ian Fellows. This package does not require GUI or Java, and intended for command-line users.
The code being executed can be found in the introduction vignette.
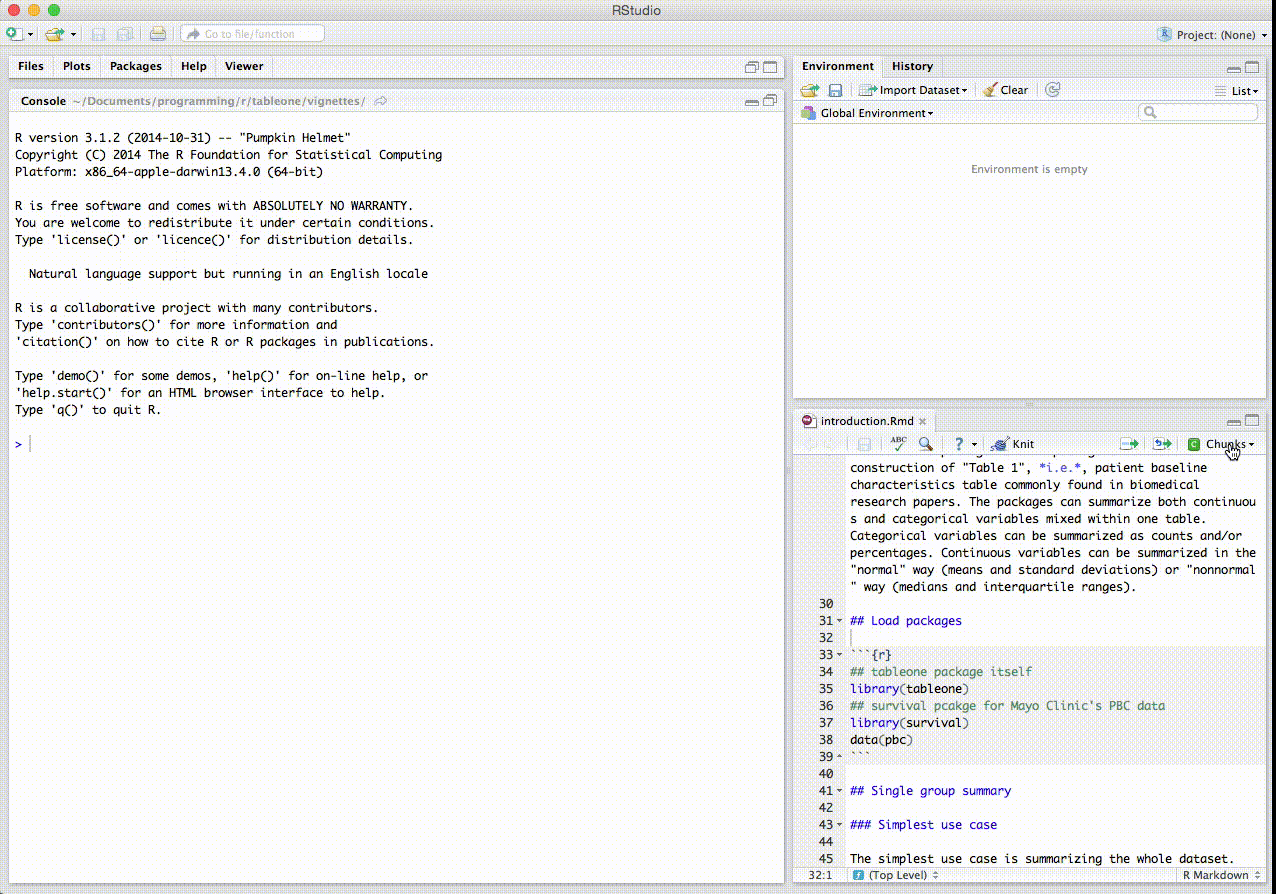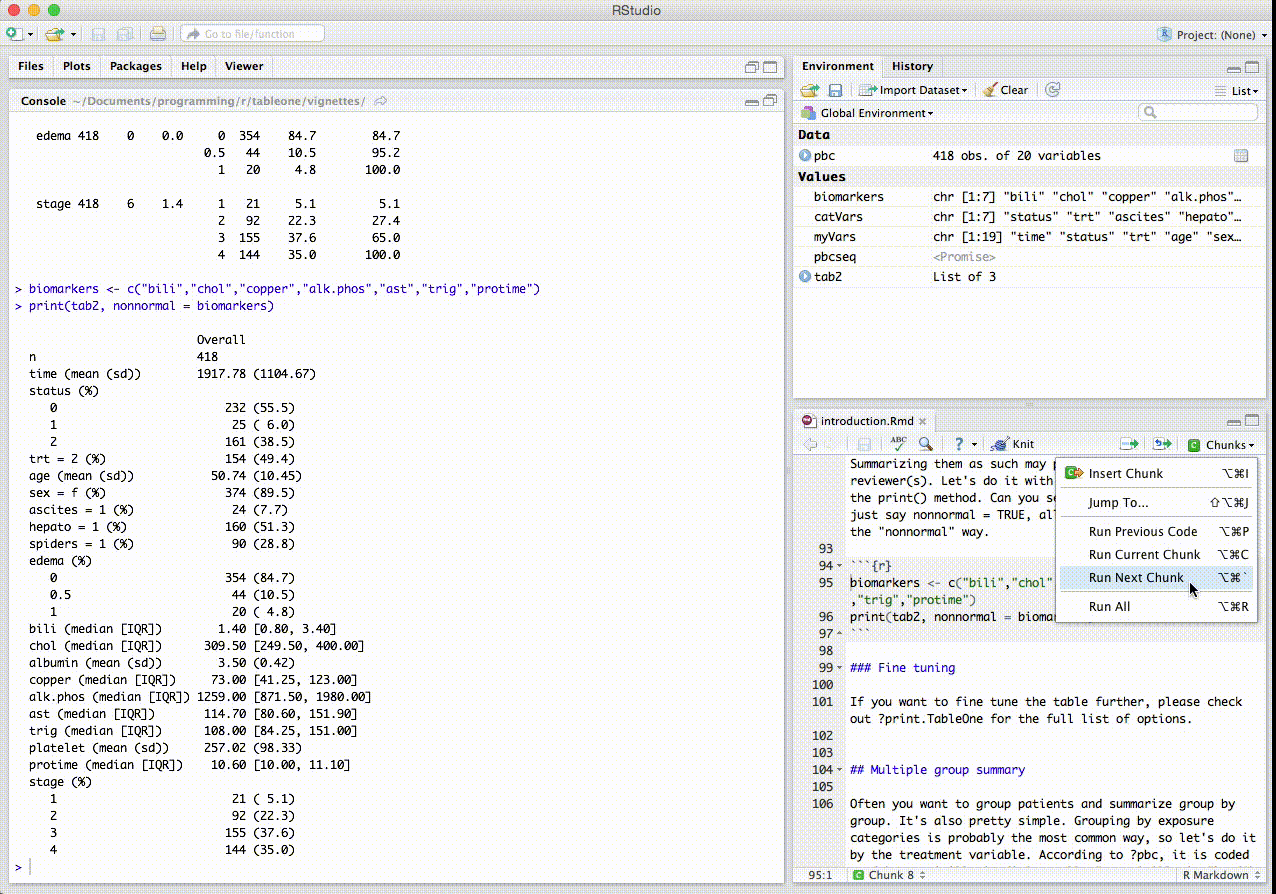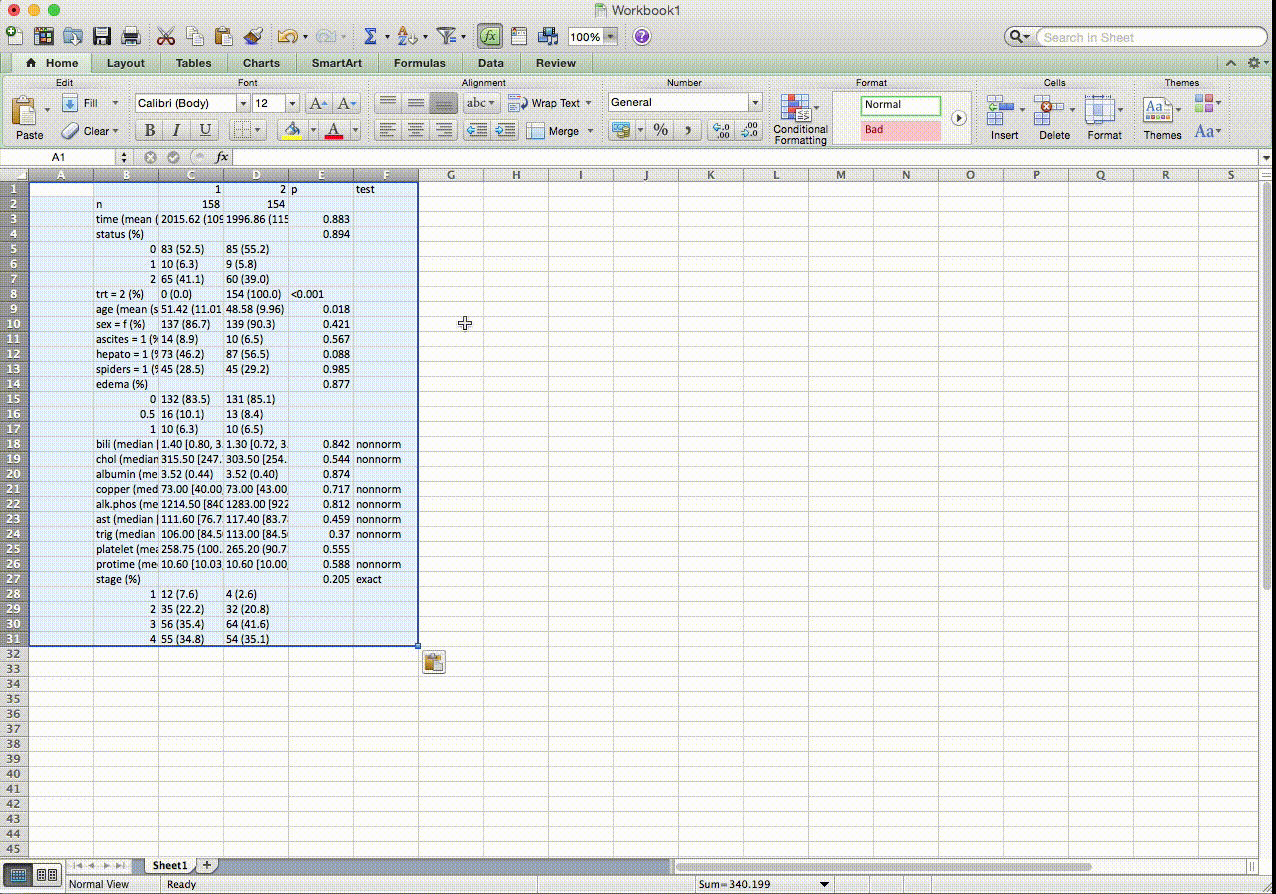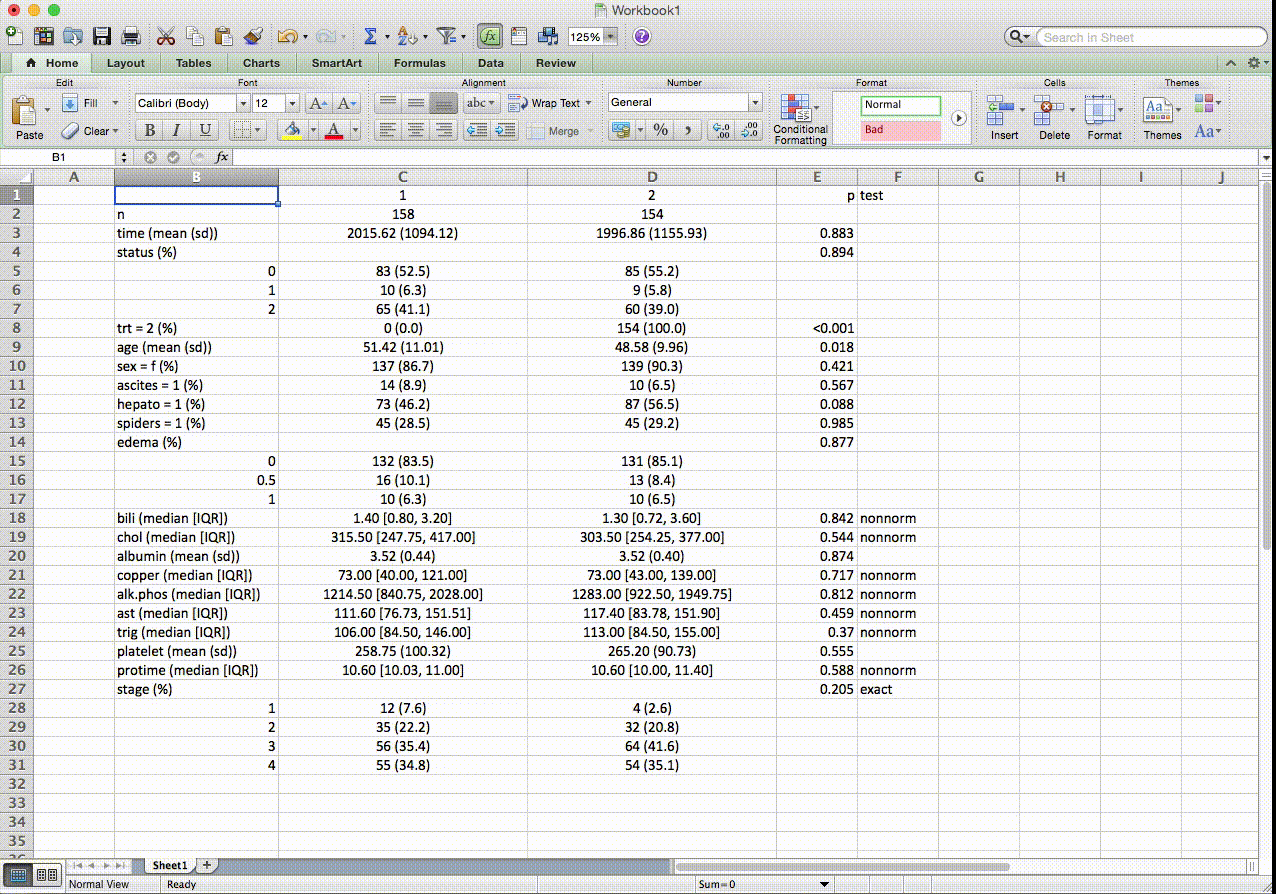
In this table, continuous and categorical variables can be placed in any order. The p-valeus are from exact tests for pre-specified variables. For nonnormal variables, it shows median and IQR instead of mean and SD, and p-values are from nonparametric tests. Numerically coded categorical variables can be transformed on the fly with factorVars. SMD stands for standardized mean differences. For weighted data, first created a svydesign object, and use the svyCreateTableOne() function. Most other options remain the same.
## Load package
library(tableone)
## Load data
data(pbc, package = "survival")
# drop ID from variable list
vars <- names(pbc)[-1]
## Create Table 1 stratified by trt (can add more stratifying variables)
tableOne <- CreateTableOne(vars = vars, strata = c("trt"), data = pbc,
factorVars = c("status","edema","stage"))
## Specifying nonnormal variables will show the variables appropriately,
## and show nonparametric test p-values. Specify variables in the exact
## argument to obtain the exact test p-values.
print(tableOne, nonnormal = c("bili","chol","copper","alk.phos","trig"),
exact = c("status","stage"), smd = TRUE,
formatOptions = list(big.mark = ","))
## Stratified by trt
## 1 2 p test SMD
## n 158 154
## time (mean (SD)) 2,015.62 (1,094.12) 1,996.86 (1,155.93) 0.883 0.017
## status (%) 0.884 exact 0.054
## 0 83 (52.5) 85 (55.2)
## 1 10 ( 6.3) 9 ( 5.8)
## 2 65 (41.1) 60 (39.0)
## trt (mean (SD)) 1.00 (0.00) 2.00 (0.00) <0.001 Inf
## age (mean (SD)) 51.42 (11.01) 48.58 (9.96) 0.018 0.270
## sex = f (%) 137 (86.7) 139 (90.3) 0.421 0.111
## ascites (mean (SD)) 0.09 (0.29) 0.06 (0.25) 0.434 0.089
## hepato (mean (SD)) 0.46 (0.50) 0.56 (0.50) 0.069 0.206
## spiders (mean (SD)) 0.28 (0.45) 0.29 (0.46) 0.886 0.016
## edema (%) 0.877 0.058
## 0 132 (83.5) 131 (85.1)
## 0.5 16 (10.1) 13 ( 8.4)
## 1 10 ( 6.3) 10 ( 6.5)
## bili (median [IQR]) 1.40 [0.80, 3.20] 1.30 [0.72, 3.60] 0.842 nonnorm 0.171
## chol (median [IQR]) 315.50 [247.75, 417.00] 303.50 [254.25, 377.00] 0.544 nonnorm 0.038
## albumin (mean (SD)) 3.52 (0.44) 3.52 (0.40) 0.874 0.018
## copper (median [IQR]) 73.00 [40.00, 121.00] 73.00 [43.00, 139.00] 0.717 nonnorm <0.001
## alk.phos (median [IQR]) 1,214.50 [840.75, 2,028.00] 1,283.00 [922.50, 1,949.75] 0.812 nonnorm 0.037
## ast (mean (SD)) 120.21 (54.52) 124.97 (58.93) 0.460 0.084
## trig (median [IQR]) 106.00 [84.50, 146.00] 113.00 [84.50, 155.00] 0.370 nonnorm 0.017
## platelet (mean (SD)) 258.75 (100.32) 265.20 (90.73) 0.555 0.067
## protime (mean (SD)) 10.65 (0.85) 10.80 (1.14) 0.197 0.146
## stage (%) 0.205 exact 0.246
## 1 12 ( 7.6) 4 ( 2.6)
## 2 35 (22.2) 32 (20.8)
## 3 56 (35.4) 64 (41.6)
## 4 55 (34.8) 54 (35.1)
This version of tableone package for R is developmetal, and may not be available from the CRAN. You can install it using one of the following way.
Direct installation from github
You first need to install the devtools package to do the following. You can choose from the latest stable version and the latest development version.
## Install devtools (if you do not have it already)
install.packages("devtools")
## Install directly from github (develop branch)
devtools::install_github(repo = "kaz-yos/tableone", ref = "develop")
Using devtools may requires some preparation, please see the following link for information.
http://www.rstudio.com/projects/devtools/
I would like to thank all the contributors!
- Alexander Bartel ndevln
- Jonathan J Chipman chipmanj
- Justin Bohn jmb01
- Lucy D’Agostino McGowan LucyMcGowan
- Malcolm Barrett malcolmbarrett
- Rune Haubo B Christensen runehaubo
- gbouzill
There are multiple similar or complementary projects of interest.
- DescTools: Tools for Descriptive Statistics. https://cran.r-project.org/web/packages/DescTools/index.html
- Gmisc: Descriptive Statistics, Transition Plots, and More. https://cran.r-project.org/web/packages/Gmisc/
- Hmisc (summary.formula): Advanced table making and many more. http://biostat.mc.vanderbilt.edu/wiki/Main/Hmisc
- arsenal: An Arsenal of ‘R’ Functions for Large-Scale Statistical Summaries. https://github.com/eheinzen/arsenal
- atable: Create Tables for Reporting Clinical Trials. https://github.com/arminstroebel/atable
- compareGroups: Descriptive Analysis by Groups. http://www.comparegroups.eu
- expss: Tables with Labels and Some Useful Functions from Spreadsheets and ‘SPSS’ Statistics. https://github.com/gdemin/expss
- finalfit: Quickly Create Elegant Regression Results Tables and Plots when Modelling. https://finalfit.org/index.html
- framework for easily create tables for reporting: framework for easily create tables for reporting. https://davidgohel.github.io/flextable/
- furniture: Furniture for Quantitative Scientists. https://cran.r-project.org/web/packages/furniture/
- gtsummary: Presentation-Ready Data Summary and Analytic Result Tables. https://CRAN.R-project.org/package=gtsummary
- htmlTable: An R package for generating advanced tables. https://github.com/gforge/htmlTable
- kableExtra: Construct Complex Table with ‘kable’ and Pipe Syntax. https://github.com/haozhu233/kableExtra
- pander: An R Pandoc Writer. http://rapporter.github.io/pander/
- pixiedust: Format models for console and to markdown, HTML, and LaTeX. https://github.com/nutterb/pixiedust
- qwraps2: quickly placing data summaries and formatted regression results into .Rnw or .Rmd files. https://github.com/dewittpe/qwraps2/
- stargazer: Well-Formatted Regression and Summary Statistics Tables. https://cran.r-project.org/web/packages/stargazer/index.html
- tab: Functions for Creating Summary Tables for Statistical Reports. https://cran.r-project.org/package=tab
- table1: Tables of Descriptive Statistics in HTML. https://github.com/benjaminrich/table1
- table1xls: Exports Reproducible Summary Tables to Multi-Tab Spreadsheet Files. https://cran.r-project.org/web/packages/table1xls/index.html
- xtable: Export Tables to LaTeX or HTML. https://cran.r-project.org/web/packages/xtable/index.html
- (Python) tableone: Create “Table 1” for research papers in Python. https://github.com/tompollard/tableone