This is a Python script to convert the output from many profilers into a dot graph.
It has the following features:
- reads output from:
- prunes nodes and edges below a certain threshold;
- uses an heuristic to propagate time inside mutually recursive functions;
- uses color efficiently to draw attention to hot-spots;
- works on any platform where Python and graphviz is available, i.e, virtually anywhere.
If you want an interactive viewer for gprof2dot output graphs, check xdot.py.
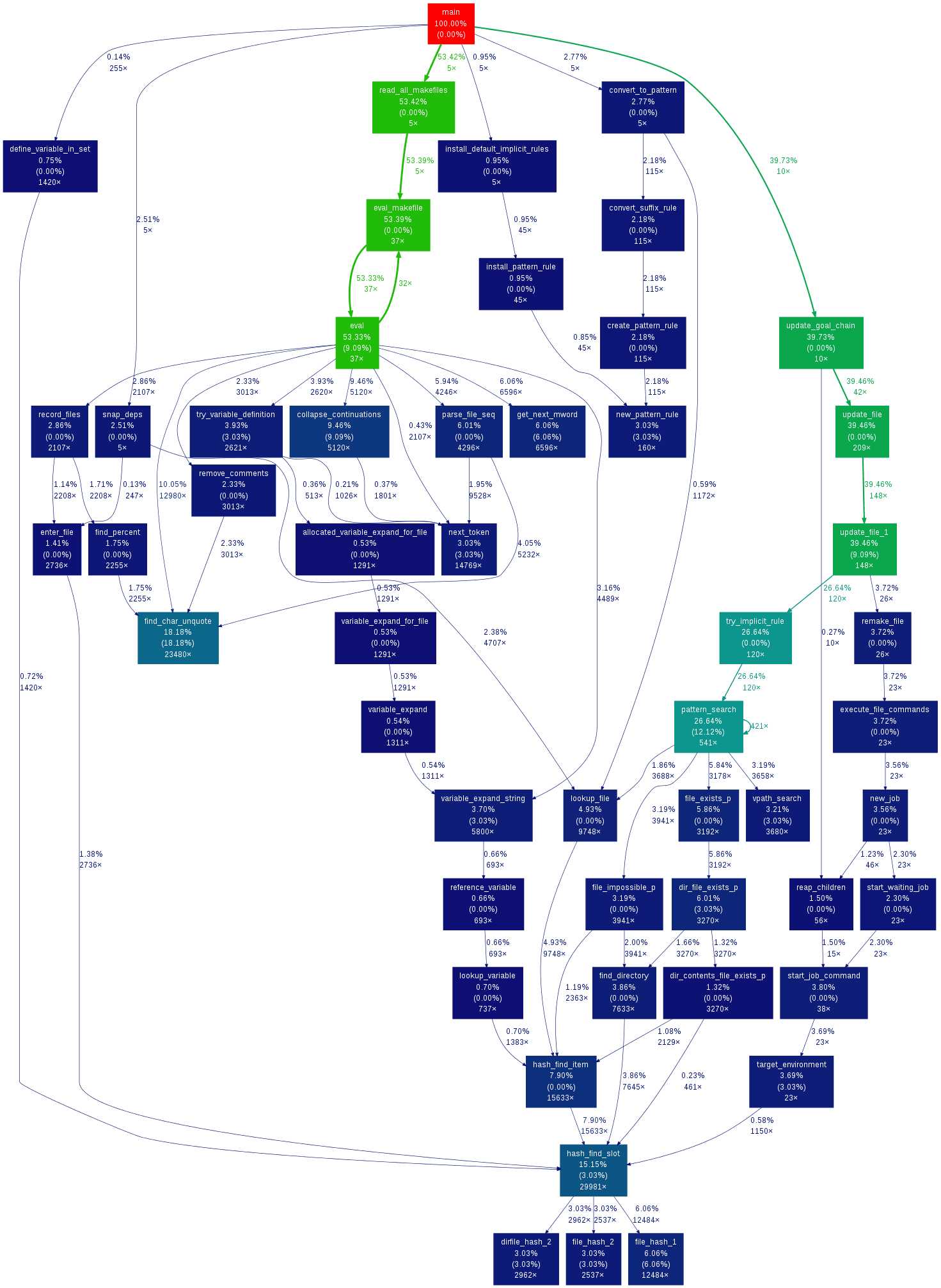
This is the result from the example data in the Linux Gazette article with the default settings:
- Python: known to work with version 2.7 and 3.3; it will most likely not work with earlier releases.
- Graphviz: tested with version 2.26.3, but should work fine with other versions.
- Download and install Python for Windows
- Download and install Graphviz for Windows
-
Run:
apt-get install python graphviz
-
pip install gprof2dot
Usage:
gprof2dot.py [options] [file] ...
Options:
-h, --help show this help message and exit
-o FILE, --output=FILE
output filename [stdout]
-n PERCENTAGE, --node-thres=PERCENTAGE
eliminate nodes below this threshold [default: 0.5]
-e PERCENTAGE, --edge-thres=PERCENTAGE
eliminate edges below this threshold [default: 0.1]
-f FORMAT, --format=FORMAT
profile format: axe, callgrind, hprof, json, oprofile,
perf, prof, pstats, sleepy, sysprof or xperf [default:
prof]
--total=TOTALMETHOD preferred method of calculating total time: callratios
or callstacks (currently affects only perf format)
[default: callratios]
-c THEME, --colormap=THEME
color map: color, pink, gray, bw, or print [default:
color]
-s, --strip strip function parameters, template parameters, and
const modifiers from demangled C++ function names
-w, --wrap wrap function names
--show-samples show function samples
-z ROOT, --root=ROOT prune call graph to show only descendants of specified
root function
-l LEAF, --leaf=LEAF prune call graph to show only ancestors of specified
leaf function
--skew=THEME_SKEW skew the colorization curve. Values < 1.0 give more
variety to lower percentages. Values > 1.0 give less
variety to lower percentages
perf record -g -- /path/to/your/executable
perf script | gprof2dot.py -f perf | dot -Tpng -o output.png
opcontrol --callgraph=16
opcontrol --start
/path/to/your/executable arg1 arg2
opcontrol --stop
opcontrol --dump
opreport -cgf | gprof2dot.py -f oprofile | dot -Tpng -o output.png
If you're not familiar with xperf then read this excellent article first. Then do:
-
Start xperf as
xperf -on Latency -stackwalk profile -
Run your application.
-
Save the data. ` xperf -d output.etl
-
Start the visualizer:
xperf output.etl -
In Trace menu, select Load Symbols. Configure Symbol Paths if necessary.
-
Select an area of interest on the CPU sampling graph, right-click, and select Summary Table.
-
In the Columns menu, make sure the Stack column is enabled and visible.
-
Right click on a row, choose Export Full Table, and save to output.csv.
-
Then invoke gprof2dot as
gprof2dot.py -f xperf output.csv | dot -Tpng -o output.png
-
Collect profile data as (also can be done from GUI):
amplxe-cl -collect hotspots -result-dir output -- your-app -
Visualize profile data as:
amplxe-cl -report gprof-cc -result-dir output -format text -report-output output.txt gprof2dot.py -f axe output.txt | dot -Tpng -o output.png
See also Kirill Rogozhin's blog post.
/path/to/your/executable arg1 arg2
gprof path/to/your/executable | gprof2dot.py | dot -Tpng -o output.png
python -m profile -o output.pstats path/to/your/script arg1 arg2
gprof2dot.py -f pstats output.pstats | dot -Tpng -o output.png
python -m cProfile -o output.pstats path/to/your/script arg1 arg2
gprof2dot.py -f pstats output.pstats | dot -Tpng -o output.png
The hotshot profiler does not include a main function. Use the hotshotmain.py script instead.
hotshotmain.py -o output.pstats path/to/your/script arg1 arg2
gprof2dot.py -f pstats output.pstats | dot -Tpng -o output.png
See Russell Power's blog post for details.
A node in the output graph represents a function and has the following layout:
+------------------------------+
| function name |
| total time % ( self time % ) |
| total calls |
+------------------------------+
where:
- total time % is the percentage of the running time spent in this function and all its children;
- self time % is the percentage of the running time spent in this function alone;
- total calls is the total number of times this function was called (including recursive calls).
An edge represents the calls between two functions and has the following layout:
total time %
calls
parent --------------------> children
Where:
- total time % is the percentage of the running time transfered from the children to this parent (if available);
- calls is the number of calls the parent function called the children.
Note that in recursive cycles, the total time % in the node is the same for the whole functions in the cycle, and there is no total time % figure in the edges inside the cycle, since such figure would make no sense.
The color of the nodes and edges varies according to the total time % value. In the default temperature-like color-map, functions where most time is spent (hot-spots) are marked as saturated red, and functions where little time is spent are marked as dark blue. Note that functions where negligible or no time is spent do not appear in the graph by default.
By default gprof2dot.py generates a partial call graph, excluding nodes and edges with little or no impact in the total computation time. If you want the full call graph then set a zero threshold for nodes and edges via the -n / --node-thres and -e / --edge-thres options, as:
gprof2dot.py -n0 -e0
The node labels can get very wide when profiling C++ code, due to inclusion of scope, function arguments, and template arguments in demangled C++ function names.
If you do not need function and template arguments information, then pass the -s / --strip option to strip them.
If you want to keep all that information, or if the labels are still too wide, then you can pass the -w / --wrap, to wrap the labels. Note that because dot does not wrap labels automatically the label margins will not be perfectly aligned.
Likely, the total execution time is too short, so there is not enough precision in the profile to determine where time is being spent.
You can still force displaying the whole graph by setting a zero threshold for nodes and edges via the -n / --node-thres and -e / --edge-thres options, as:
gprof2dot.py -n0 -e0
But to get meaningful results you will need to find a way to run the program for a longer time period (aggregate results from multiple runs).
You likely have an execution time too short, causing the round-off errors to be large.
See question above for ways to increase execution time.
Options which are essential to produce suitable results are:
-g: produce debugging information-fno-omit-frame-pointer: use the frame pointer (frame pointer usage is disabled by default in some architectures like x86_64 and for some optimization levels; it is impossible to walk the call stack without it)
If you're using gprof you will also need -pg option, but nowadays you can get much better results with other profiling tools, most of which require no special code instrumentation when compiling.
You want the code you are profiling to be as close as possible as the code that you will be releasing. So you should include all options that you use in your release code, typically:
-O2: optimizations that do not involve a space-speed tradeoff-DNDEBUG: disable debugging code in the standard library (such as the assert macro)
However many of the optimizations performed by gcc interfere with the accuracy/granularity of the profiling results. You should pass these options to disable those particular optimizations:
-fno-inline-functions: do not inline functions into their parents (otherwise the time spent on these functions will be attributed to the caller)-fno-inline-functions-called-once: similar to above-fno-optimize-sibling-calls: do not optimize sibling and tail recursive calls (otherwise tail calls may be attributed to the parent function)
If the granularity is still too low, you may pass these options to achieve finer granularity:
-fno-default-inline: do not make member functions inline by default merely because they are defined inside the class scope-fno-inline: do not pay attention to the inline keyword Note however that with these last options the timings of functions called many times will be distorted due to the function call overhead. This is particularly true for typical C++ code which expects that these optimizations to be done for decent performance.
See the full list of gcc optimization options for more information.
See the wiki for external resources, including complementary/alternative tools.