As we know, OpenFOAM combines all species to a mixture by the mass fraction weighted average.
But for thermo.dat, we have Tlow_ Thigh_ Tcommon_.
During the average procedure, operator+= is called.
You will find that the final mixture_ will get the lowest Tlow_, highest Thigh_.
However, Tcommon_ is never changed, which means mixture_ will be that of the first species.
Usually, Tcommon_ is 1000 K, but it's a different story for some species, such as 1400 K.
If you, unfortunately, list the species 1400 K as the first one, as I warned here, you will face this problem after the Tmax of your CFD simulation reaches 1400 K:
Maximum number of iterations exceeded
It is because the Newton iterations failed when calculating temperature from enthalpy, and the discontinuous cp is the key to the problem.
And this exactly attributes to the wrong Tcommon.
The simplest but not perfect solution is to put the 1000 K species as the first one (in chem.inp, where OpenFOAM reads species list).
Also, you can use fitData utility in Chemkin, which can only fit one species at a time.
This script is developed to completely solve this problem. Only need to run once this script, you'll get a fresh new thermo.dat, with a uniform Tcommon, which will give you continuous cp.
- install Cantera
conda create --name spam --channel cantera cantera ipython matplotlib- activate Cantera
conda activate spam- transform your chemkin format mechanism to
*.ctiformat
ck2cti --input=chem.inp --thermo=thermo.dat --output=mech.cti- run the script
python fitData_Cantera.py- you will get thermoCorrected.dat
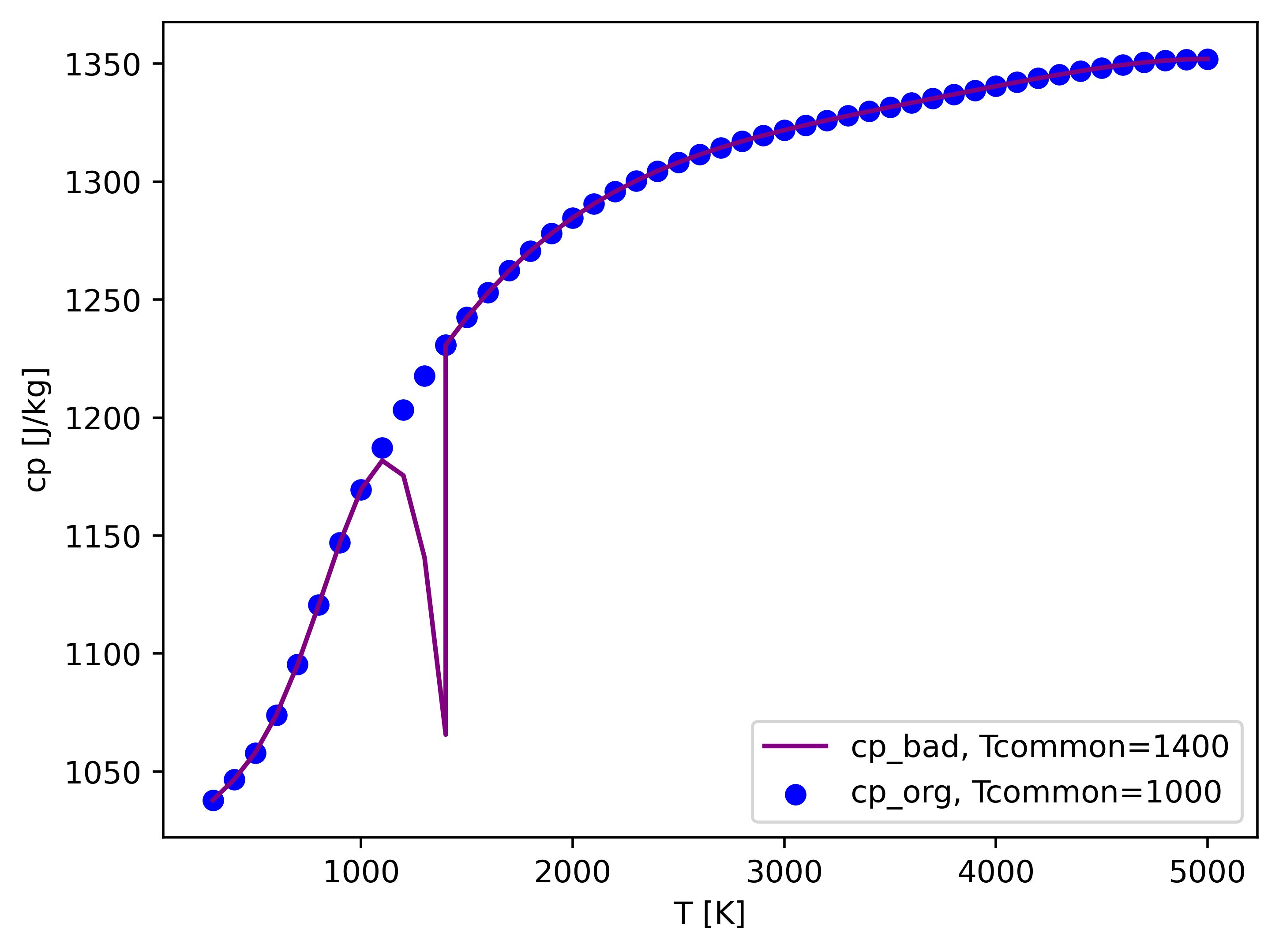
Here is an example of the plotted figure.
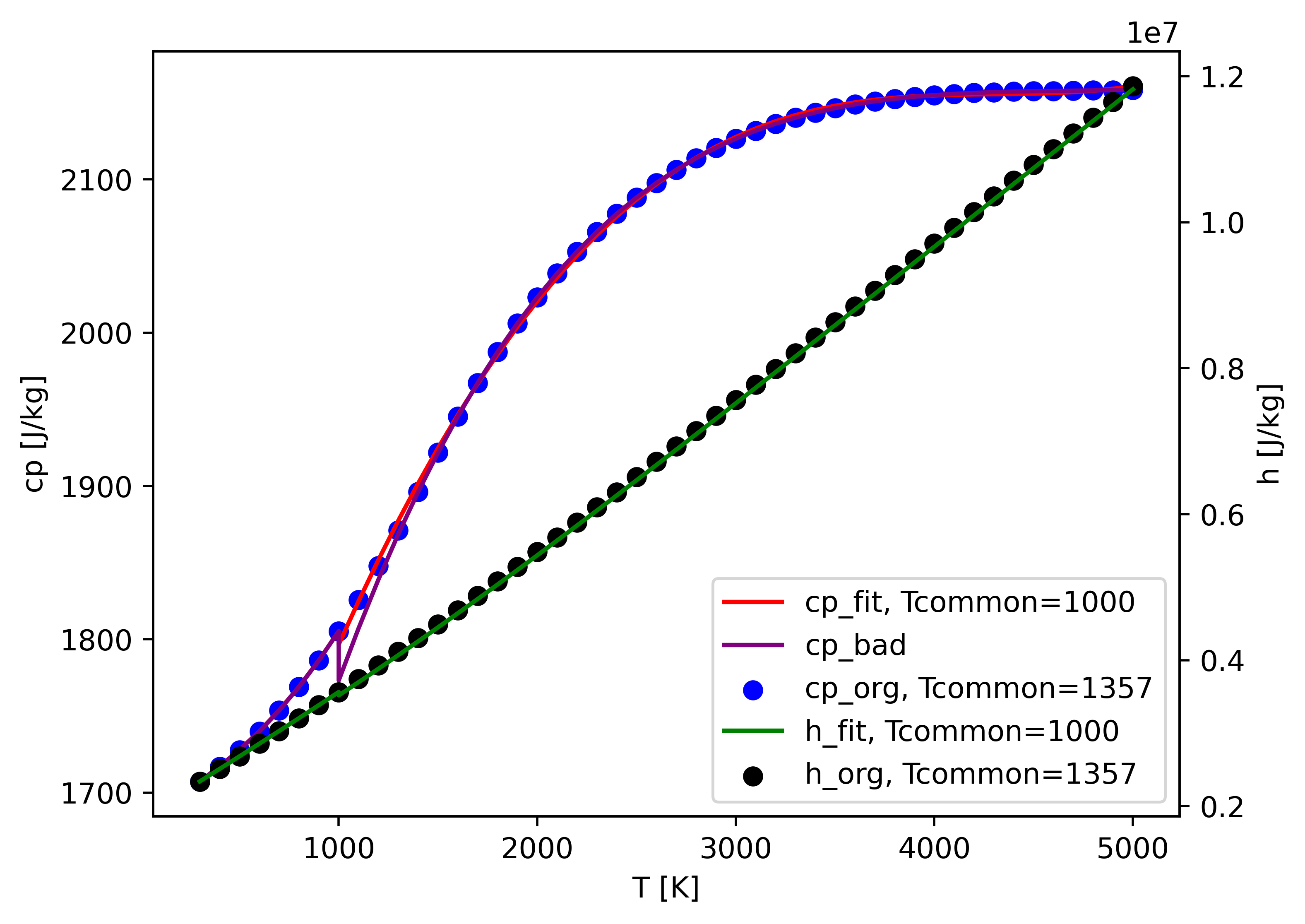 For OH, we fit the Tcommon from 1357 to 1000 K.
For OH, we fit the Tcommon from 1357 to 1000 K.
cp_org is calculated by Cantera using the original mechanism file, which is set as the correct answer.
cp_fit is calculated using the newly fitted coeffs with new Tcommon.
And if you just manually change 1357 to 1000 in thermo.dat. You'll get the purple cp_bad, with a discontinuous cp.
However, this is not the worst condition! And your case may still run normally, but with little error introduced.
If you mistakenly set a species with 'Tcommon = 1400 K' as the first species in 'chem.inp' (used in OpenFOAM), you will most likely get a wrong answer if the original Tcommon of N2 is 1000 K (here we assume N2 is the majority species).