This is Ligo's open-source implementation of AlphaFold3, an ongoing research project aimed at advancing open-source biomolecular structure prediction. This release implements the full AlphaFold3 model along with the training code. We are releasing the single chain prediction capability first and we will add ligand, multimer, and nucleic acid prediction capabilities once they are trained. Sign up for beta testing here.
This repository is intended to accelerate progress towards a faithful, fully open-source implementation of AlphaFold3 for the entire biotech community to use freely.
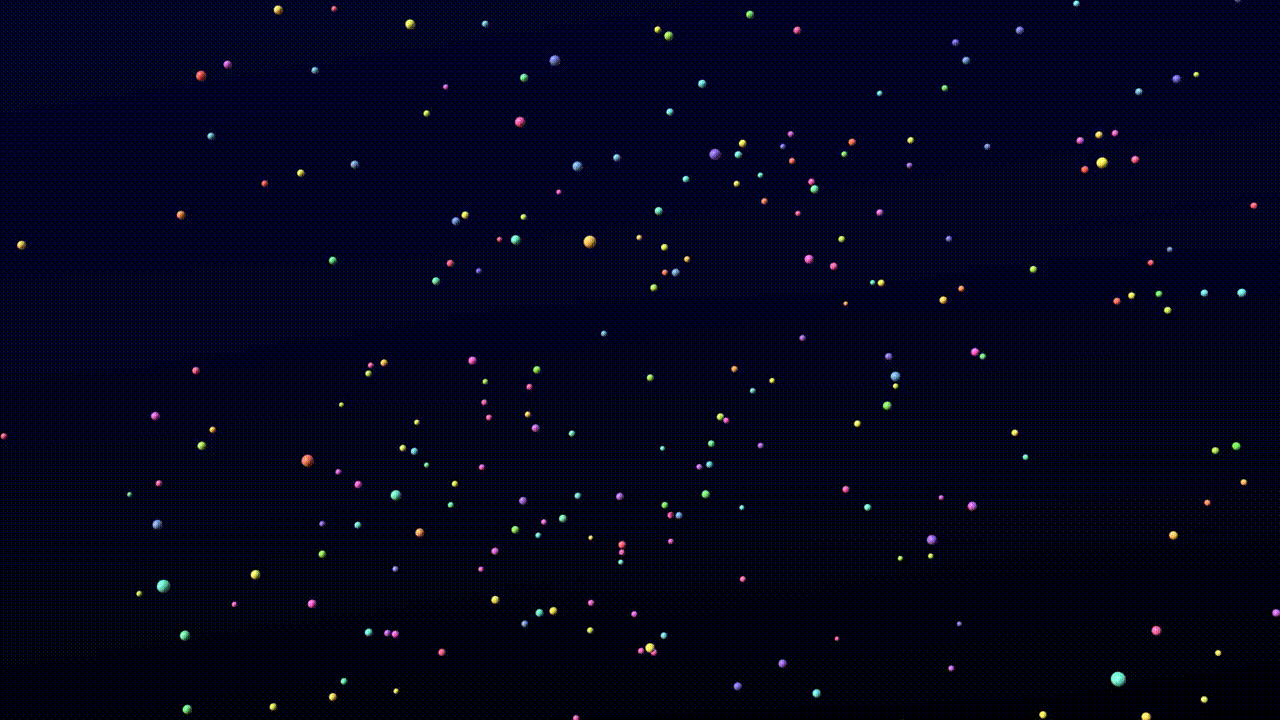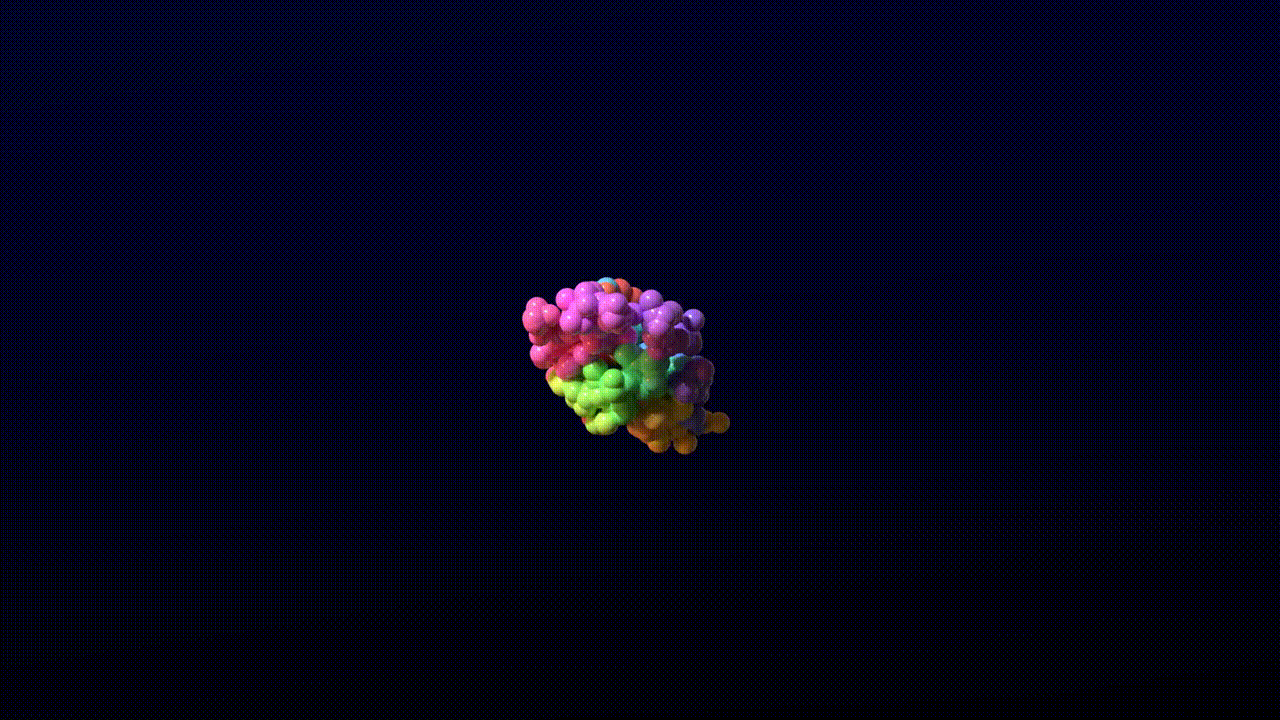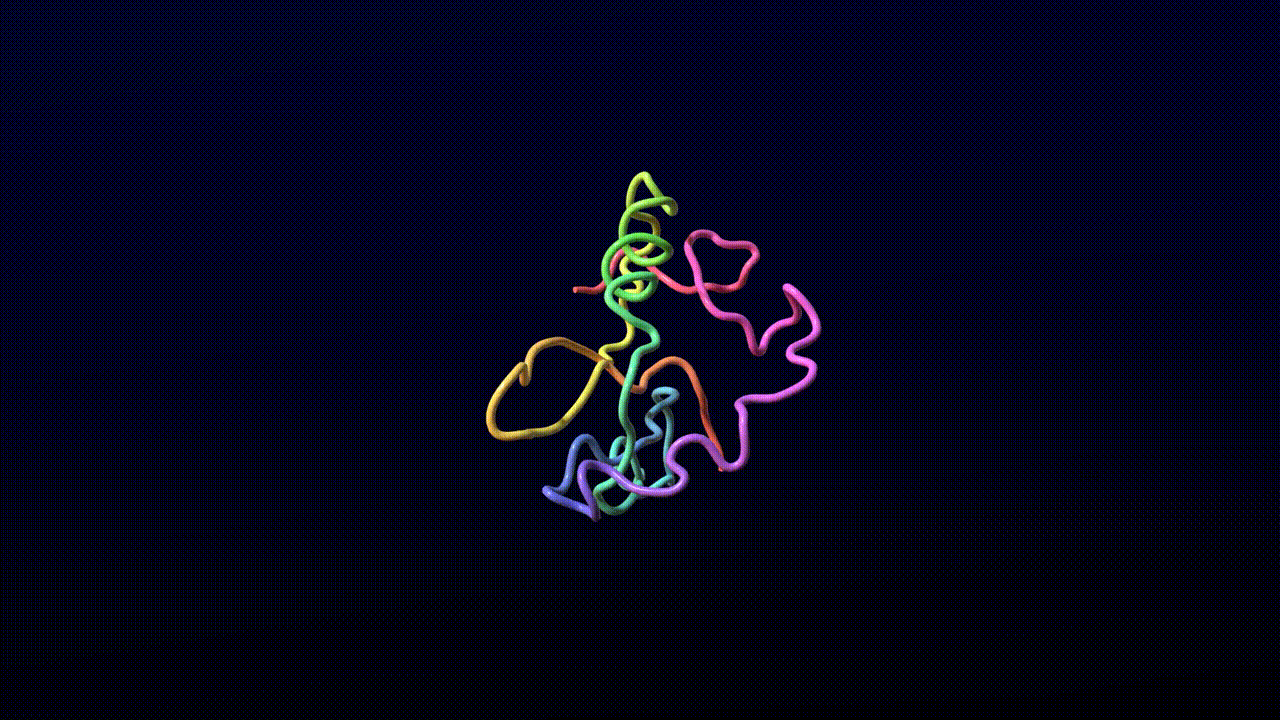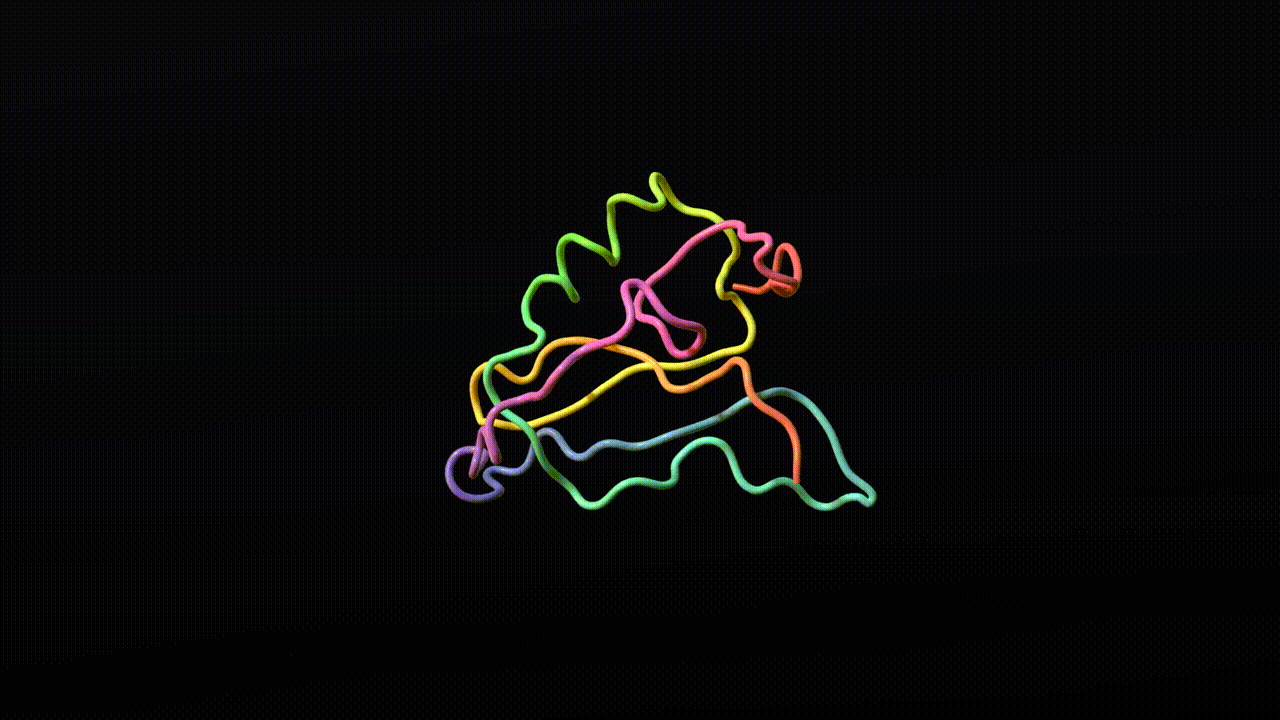
We find that the model training dynamics are quite fast. The following video is a sample from a model trained for 4,000 steps on 8 A100 GPUs for 10 hours without templates.
Animation credits: Matthew Clark
This project would not have been possible without the contributions of the following projects and individuals:
-
The AlphaFold3 team at Google DeepMind for their groundbreaking work and publishing the core algorithms.
-
The OpenFold project (https://github.com/aqlaboratory/openfold), which laid the foundation for open-source protein structure prediction. We reuse many of their core modules, such as triangular attention and multiplicative update, as well as their data processing pipelines.
-
The ProteinFlow library (https://github.com/adaptyvbio/ProteinFlow), especially the architect of ProteinFlow, Liza Kozlova (@elkoz), who has been an absolute hero throughout this process. We trained most of our prototype models on ProteinFlow, since it provides a clean and well-documented data pipeline for working with protein data. We have partnered with AdaptyvBio to build the data pipeline of AlphaFold3 based on ProteinFlow that includes full ligand and nucleic acid support. @elkoz and @igor-krawczuk are building the next release of ProteinFlow to include full support for these data modalities.
This is an active research project in its early phases. We are working to prepare a stable release for the community. While we are excited about the potential of this work, we want to emphasise that this is not yet a production-ready tool. We trained a version of AlphaFold3 on single-chain proteins to test the implementation -- the next release will include full ligand and nucleic acid support. We are accepting a small number of beta testers to help us test the implementation and provide feedback. If you are interested in beta testing, please join our waitlist.
While working on this project, we discovered a few properties of the algorithms described in the AlphaFold3 supplementary information that were not consistent with surrounding deep learning literature. We discuss these below:
-
MSA Module Order: In the Supplementary Information, the MSA module communication step occurs before the MSA stack. This results in the MSA stack of the last block not contributing to the structure prediction, since all information flows out through the pair representation. With the order in the pseudocode, the MSA stack in the last block does not have an opportunity to update the pair representation. We swap the OuterProductMean operation and the MSA stack to ensure all blocks contribute to the structure prediction. It is important to note this correction is consistent with the order of operations in the ExtraMSAStack of AlphaFold2. DeepMind mentions these MSA module blocks are "homogeneous". It is unclear whether this means shared weights or same architecture across blocks. If the layers are shared, then gradients will flow through all of them but the final calculation of the MSA stack is idle - this can be safely skipped (not mentioned in the pseudocode). We will resolve this ambiguity in light of DeepMind's response.
-
Loss scaling: The loss scaling factor described in the Supplementary Information does not give unit-loss at initialization. Unit-loss at initialization is one of the properties that Karras et al. (2022) set as a desirable property of the loss function when training diffusion models, and Max Jaderberg mentions this as one of the properties for why they chose the framework of Karras et al. in this talk here. We think this is a simple typo in the Supplementary info that is due to an addition being typed as a multiplication -- in our implementation, we use the loss scaling factor consistent with Karras et al. (2022). Our measurements show that this gives unit MSE loss at initialization, whilst the scaling in the Supplementary Information is two to three orders of magnitude larger at initialization. Additionally, the loss scaling factor in the paper has a local minimum at t = 16.0, but then it increases with increasing noise level. This is not in line with the properties of the loss function that Karras et al. (2022) proposed, which emphasises the importance of downweighting the loss at higher noise levels. We add a Jupyter notebook to the repository showing our experiments.
-
DiT block design: The design of the AttentionPairBias and the DiffusionTransformer blocks seem to closely follow the DiT block design introduced by Peebles & Xie (2022) here. However, the residual connections are missing. It is not explained in the paper why DeepMind chose to omit them. We experiment with both and find that (within the range of steps we trained our models on) the DiT block with residual connections gives much faster convergence and better gradient flow through the network. Note that this is the discrepancy we are the least sure about, and it can be changed in a couple lines in our code if the original implementation does not use the residual connections.
These are noted here for transparency and to invite community input on the best approaches to resolve them.
A significant focus of this implementation has been on optimising the model components for speed and memory efficiency. AlphaFold3 has many transformer-like components, but efficient hardware-aware attention implementations like FlashAttention2 do not integrate out-of-the-box with these modules due to pair biasing in AlphaFold3. All of the attention operations project a pair bias from the pairwise representation that is added after the key-query dot product, and the bias requires a gradient to be backpropagated. This is not out of scope for FlashAttention2, since the bias gradient would have the same gradient as the scaled QK^T dot product, but the current implementation does not support this. More recent attention implementations like FlexAttention are very promising, but they also do not support a bias gradient for now since broadcasting operations of the bias tensor during the forward pass become reductions in the backward pass, and this functionality is not implemented in the first release of FlexAttention.
-
We reuse battle-tested components such as TriangularAttention and TriangularMultiplicativeUpdate from the OpenFold project wherever we can. The modular design of the OpenFold project allows us to easily import these modules into our codebase. We are working on improving the efficiency of these modules with Triton, fusing operations to increase performance and reduce intermediate tensor allocation.
-
We observed that a naive implementation of the Diffusion Module in PyTorch frequently ran out of memory since the Diffusion Module is replicated 48 times per batch. To solve this issue, we re-purpose the MSARowAttentionWithPairBias kernel from Deepspeed4Science to implement a memory-efficient version of the Diffusion Module, treating the batch replicas with different noise levels as an additional batch dimension. For the AtomAttentionEncoder and AtomAttentionDecoder modules, we experimented with a custom PyTorch-native implementation to reduce the memory footprint from quadratic to linear, but the benefits were not that significant compared to a naive re-purposing of the AttentionPairBias kernel. We include both implementations in the repository, but use the naive implementation for the sake of reducing clutter. Despite these optimisations, our profiling experiments show that over 60% of the model's operations are memory-bound. We are working on a far more efficient and scalable implementation using the ideas of ScaleFold, which will allow us to reach the training scale of the original AlphaFold3.
We do not yet provide sampling code since the ligand-protein and nucleic acid prediction capabilities are yet to be trained. The checkpoint weights can be loaded with PyTorch Lightning's checkpoint loading for experimentation and model surgery. The current model only predicts single-chain proteins, which is the same functionality as the original AlphaFold2. The model components are written to be reusable and modular so that researchers can easily incorporate them into their own projects. For beta testing of ligand-protein and nucleic acid prediction: Join our Waitlist
For now, the primary use of this repository is for research and development. We will include more user-facing functionality in the future once the ligand-protein and nucleic acid prediction capabilities are ready.
We welcome contributions from the community! There are likely numerous bugs and subtle implementation errors in our code. Deep learning training often fails silently, where the errors still allow the network to converge but make it work slightly worse. If you're interested in contributing, you can raise a Github issue with a bug description or fork the repository, create a new branch with your corrections and submit a pull request with a clear description of your changes.
For any other comments or suggestions please contact us via email at alphafold3@ligo.bio.
If you use this code in your research, please cite the following papers:
@article{Abramson2024-fj,
title = "Accurate structure prediction of biomolecular interactions with
{AlphaFold} 3",
author = "Abramson, Josh and Adler, Jonas and Dunger, Jack and Evans,
Richard and Green, Tim and Pritzel, Alexander and Ronneberger,
Olaf and Willmore, Lindsay and Ballard, Andrew J and Bambrick,
Joshua and Bodenstein, Sebastian W and Evans, David A and Hung,
Chia-Chun and O'Neill, Michael and Reiman, David and
Tunyasuvunakool, Kathryn and Wu, Zachary and {\v Z}emgulyt{\.e},
Akvil{\.e} and Arvaniti, Eirini and Beattie, Charles and
Bertolli, Ottavia and Bridgland, Alex and Cherepanov, Alexey and
Congreve, Miles and Cowen-Rivers, Alexander I and Cowie, Andrew
and Figurnov, Michael and Fuchs, Fabian B and Gladman, Hannah and
Jain, Rishub and Khan, Yousuf A and Low, Caroline M R and Perlin,
Kuba and Potapenko, Anna and Savy, Pascal and Singh, Sukhdeep and
Stecula, Adrian and Thillaisundaram, Ashok and Tong, Catherine
and Yakneen, Sergei and Zhong, Ellen D and Zielinski, Michal and
{\v Z}{\'\i}dek, Augustin and Bapst, Victor and Kohli, Pushmeet
and Jaderberg, Max and Hassabis, Demis and Jumper, John M",
journal = "Nature",
month = "May",
year = 2024
}@article {Ahdritz2022.11.20.517210,
author = {Ahdritz, Gustaf and Bouatta, Nazim and Floristean, Christina and Kadyan, Sachin and Xia, Qinghui and Gerecke, William and O{\textquoteright}Donnell, Timothy J and Berenberg, Daniel and Fisk, Ian and Zanichelli, Niccolò and Zhang, Bo and Nowaczynski, Arkadiusz and Wang, Bei and Stepniewska-Dziubinska, Marta M and Zhang, Shang and Ojewole, Adegoke and Guney, Murat Efe and Biderman, Stella and Watkins, Andrew M and Ra, Stephen and Lorenzo, Pablo Ribalta and Nivon, Lucas and Weitzner, Brian and Ban, Yih-En Andrew and Sorger, Peter K and Mostaque, Emad and Zhang, Zhao and Bonneau, Richard and AlQuraishi, Mohammed},
title = {{O}pen{F}old: {R}etraining {A}lpha{F}old2 yields new insights into its learning mechanisms and capacity for generalization},
elocation-id = {2022.11.20.517210},
year = {2022},
doi = {10.1101/2022.11.20.517210},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/10.1101/2022.11.20.517210},
eprint = {https://www.biorxiv.org/content/early/2022/11/22/2022.11.20.517210.full.pdf},
journal = {bioRxiv}
}@article{kozlova_2023_proteinflow,
author = {Kozlova, Elizaveta and Valentin, Arthur and Khadhraoui, Aous and Gutierrez, Daniel Nakhaee-Zadeh},
month = {09},
title = {ProteinFlow: a Python Library to Pre-Process Protein Structure Data for Deep Learning Applications},
doi = {https://doi.org/10.1101/2023.09.25.559346},
year = {2023},
journal = {bioRxiv}
}@misc{ahdritz2023openproteinset,
title={{O}pen{P}rotein{S}et: {T}raining data for structural biology at scale},
author={Gustaf Ahdritz and Nazim Bouatta and Sachin Kadyan and Lukas Jarosch and Daniel Berenberg and Ian Fisk and Andrew M. Watkins and Stephen Ra and Richard Bonneau and Mohammed AlQuraishi},
year={2023},
eprint={2308.05326},
archivePrefix={arXiv},
primaryClass={q-bio.BM}
}@article{Peebles2022DiT,
title={Scalable Diffusion Models with Transformers},
author={William Peebles and Saining Xie},
year={2022},
journal={arXiv preprint arXiv:2212.09748},
}@inproceedings{Karras2022edm,
author = {Tero Karras and Miika Aittala and Timo Aila and Samuli Laine},
title = {Elucidating the Design Space of Diffusion-Based Generative Models},
booktitle = {Proc. NeurIPS},
year = {2022}
}This project is licensed under the Apache License 2.0 - see the LICENSE file for details.