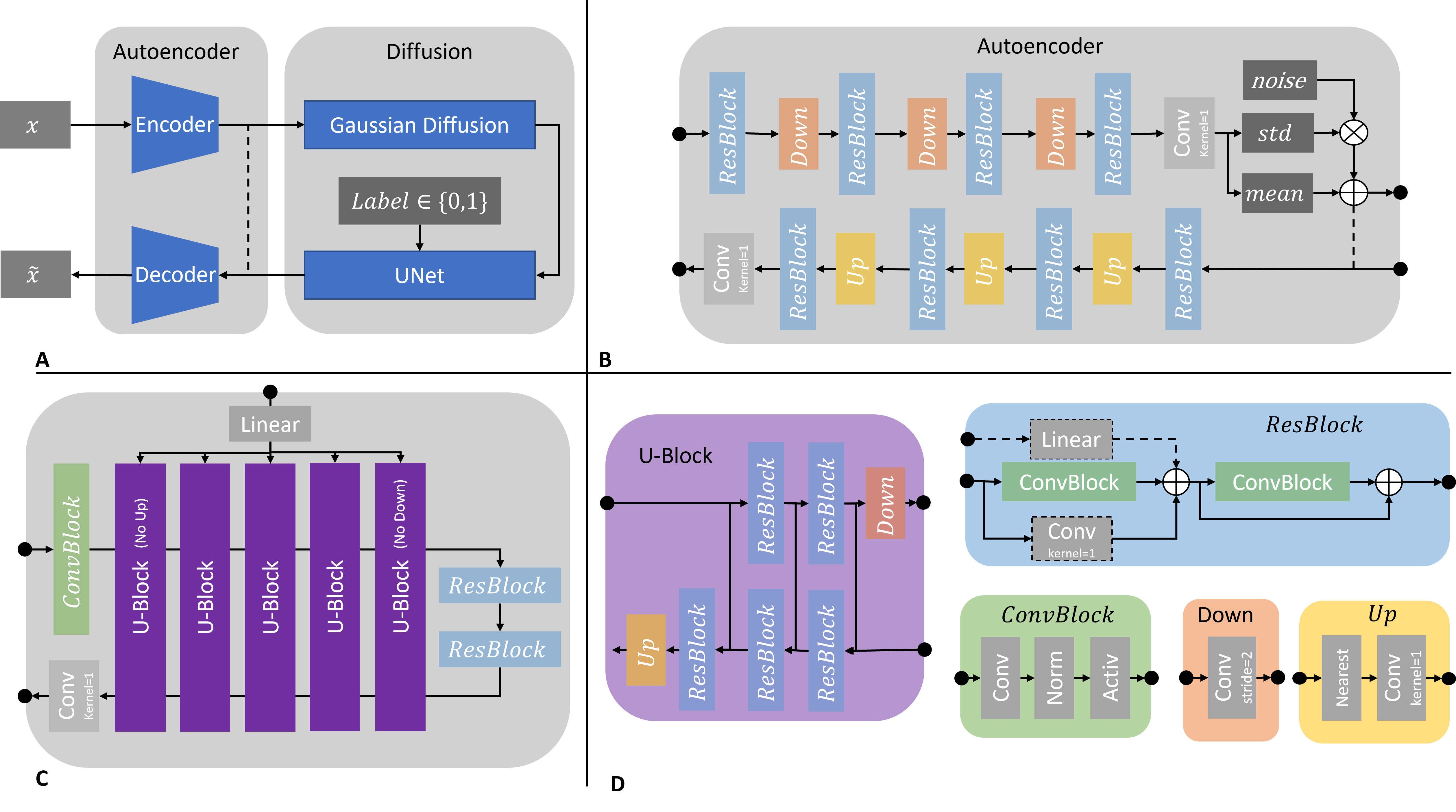
Please see: Diffusion Probabilistic Models beat GANs on Medical 2D Images
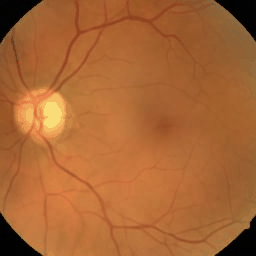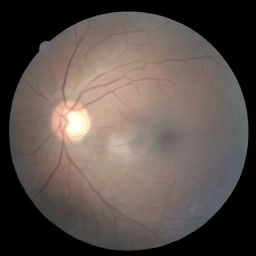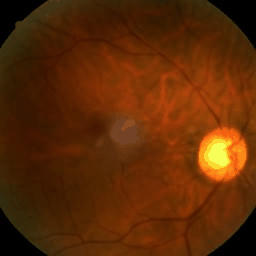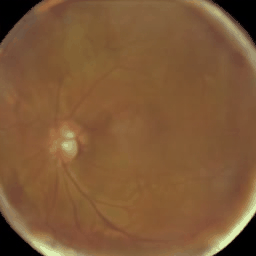


Figure: Eye fundus, chest X-ray and colon histology images generated with Medfusion (Warning color quality limited by .gif)
Link to streamlit app.
Create virtual environment and install packages:
python -m venv venv
source venv/bin/activate
pip install -e .
- Go to medical_diffusion/data/datasets/dataset_simple_2d.py and create a new
SimpleDataset2Dor write your own Dataset.
- Go to scripts/train_latent_embedder_2d.py and import your Dataset.
- Load your dataset with eg.
SimpleDataModule - Customize
VAEto your needs - (Optional): Train a
VAEGANinstead or load a pre-trainedVAEand setstart_gan_train_step=-1to start training of GAN immediately.
- Use scripts/evaluate_latent_embedder.py to evaluate the performance of the Autoencoder.
- Go to scripts/train_diffusion.py and import/load your Dataset as before.
- Load your pre-trained VAE or VAEGAN with
latent_embedder_checkpoint=... - Use
cond_embedder = LabelEmbedderfor conditional training, otherwisecond_embedder = None
- Go to scripts/sample.py to sample a test image.
- Go to scripts/helpers/sample_dataset.py to sample a more reprensative sample size.
- Use scripts/evaluate_images.py to evaluate performance of sample (FID, Precision, Recall)
- Code builds upon https://github.com/lucidrains/denoising-diffusion-pytorch