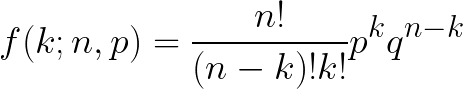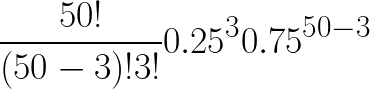The goal of this exercise is to learn some best practices for writing Python code.
At this point, you should have (1) an account on Github and (2) been introduced to the very basics of Git.
-
Login to your Github account.
-
Fork this repository, by clicking the 'Fork' button on the upper right of the page.
After a few seconds, you should be looking at your copy of the repo in your own Github account.
-
Click the 'Clone or download' button, and copy the URL of the repo via the 'copy to clipboard' button.
-
In your terminal, navigate to where you want to keep this repo (you can always move it later, so just your home directory is fine). Then type:
$ git clone the-url-you-just-copiedand hit enter to clone the repository. Make sure you are cloning your fork of this repo.
-
Next,
cdinto the directory:$ cd the-name-of-directory-you-just-cloned -
At this point, you should be in your own local copy of the repository.
-
As you work on the exercise below, be sure to frequently
addandcommityour work andpushchanges to the remote copy of the repo hosted on GitHub. Don't enter these commands now; this is just to jog your memory:$ # Do some work $ git add file-you-worked-on.py $ git commit $ git push origin master
Learn to implement recursion to a set of collected data organized using object-oriented programming techniques.
You will identify the frequency of a trait within a sampleset and determine the probability of your findings given the frequency of this allele within the population.
A recursive function calls itself one or more times until a specified condition is met, at which time the reset of each repitition is processed from the most recent call to the original.
Recursive processes are useful when the space to be examined is complex (e.g. tree traversal). While we aren't going to address these scenarios in this class, we will introduce the process of recursion using a simple example: the factorial.
In introductory math classes you learn that factorials are the product of all positive numbers from 1 to a given number n (represented as n!). For example, 5! can be written out as 5 * 4 * 3 * 2 * 1. This mathematical process can be coded iteravely as follows:
def get_factorial_iter(n):
result = 0
j = 0
for i in range(1,n+1):
result *= i
return result
However, you can also code this process recursively as follows:
def get_factorial_recur(n):
if n == 1:
return 1
else:
return n * get_factorial_recur(n-1)
A Bernoulli trial is a random experiment with two possible outcomes. Using a binomial distribution, the probability of an outcome can be calculated using the following formula:
Where k = the number of occurences of a trait, n = the sample size, p = the frequency of a given trait in a population, and q = frequency of the other trait (assuming there are only two traits). Helpful hint: p + q = 1.
Note: For the scripting for biologists class, you do not need to understand the logistics of this equation. You will simply need to know how to code it when provided the variables.
For example, if the frequency of a trait in a population is 0.25 and you collect 50 individuals from the population, here is how you would calculate the probability that 3 of the 50 individuals have that trait:
Which value is 4.11 * 10^-4.
Weagle feather color is encoded by the Aubie gene. The trait "blue" has a frequency of 0.7 within a population of weagles. This trait is encoded by the Aubie gene when the fourth amino acid is a Serine (Ser; S). The only other trait, "orange", occurs when the fourth amino acid is an Argenine (Arg;R).
Note: The Aubie gene is encoded with the vertebrate nuclear genetic code (the default genetic code for the Bio.Seq module)
Within this repository, there is a fasta file titled Aubie.fasta. This file contains a header with sample information and the coding sequence of the Aubie gene. The header contains the sample ID and the date of the sequencing, which are separated by an underscore. You are to do the following:
- Create a class with attributes for each individual's (A) sampleID, (B) date of sequencing, (C) phenotype ("orange" or "blue"), and (D) nucleotide sequence.
- Calculate the total number of individuals in your sample set (your "n")
- Provide a list of all individuals with the "orange" trait (your "k")
- Calculate the probability of finding your number of observed orange traits given the sample size and the frequency of orange within the population [f(k;n,p)]
The results of your analysis will be reported in a file called results.txt. An example is available in the file results-example.txt, which is the output from examination of the Aubie.fasta file; your results file should be in this exact format. You can use the results-example.txt file as a key as you work with the Aubie.fasta file, but your script should be able to do these calculations for any file formated the same as Aubie.fasta.
Your script should create this results.txt file from a user-provided (1) fasta file, (2) trait frequency value, and (3) output file name (results.txt). These arguments should be provided in this order from the command line when the script is ran. For example, the script to generate the results.txt would be called by running:
python popgen_bernoulli.py Aubie.fasta 0.3 results.txt
The arguments Aubie.fasta, 0.3, and results.txt are accessed by the popgen_bernoulli.py script using the sys python module.
To compare your results.txt file from analyzing the Aubie.fasta with the results-example.txt output file, you can do the following while in a directory containing both of these files:
diff results.txt results-example.txt
If this command gives you no output, there are no differences between the two files. Good job! Make sure your script fulfills the requirements described above (#s 1-5) AND that it follows best practices for python (i.e., clear variable names, functions, docstrings, modularity).